Author: Jason Brownlee
Outliers or anomalies are rare examples that do not fit in with the rest of the data.
Identifying outliers in data is referred to as outlier or anomaly detection and a subfield of machine learning focused on this problem is referred to as one-class classification. These are unsupervised learning algorithms that attempt to model “normal” examples in order to classify new examples as either normal or abnormal (e.g. outliers).
One-class classification algorithms can be used for binary classification tasks with a severely skewed class distribution. These techniques can be fit on the input examples from the majority class in the training dataset, then evaluated on a holdout test dataset.
Although not designed for these types of problems, one-class classification algorithms can be effective for imbalanced classification datasets where there are none or very few examples of the minority class, or datasets where there is no coherent structure to separate the classes that could be learned by a supervised algorithm.
In this tutorial, you will discover how to use one-class classification algorithms for datasets with severely skewed class distributions.
After completing this tutorial, you will know:
- One-class classification is a field of machine learning that provides techniques for outlier and anomaly detection.
- How to adapt one-class classification algorithms for imbalanced classification with a severely skewed class distribution.
- How to fit and evaluate one-class classification algorithms such as SVM, isolation forest, elliptic envelope, and local outlier factor.
Discover SMOTE, one-class classification, cost-sensitive learning, threshold moving, and much more in my new book, with 30 step-by-step tutorials and full Python source code.
Let’s get started.

One-Class Classification Algorithms for Imbalanced Classification
Photo by Kosala Bandara, some rights reserved.
Tutorial Overview
This tutorial is divided into five parts; they are:
- One-Class Classification for Imbalanced Data
- One-Class Support Vector Machines
- Isolation Forest
- Minimum Covariance Determinant
- Local Outlier Factor
One-Class Classification for Imbalanced Data
Outliers are both rare and unusual.
Rarity suggests that they have a low frequency relative to non-outlier data (so-called inliers). Unusual suggests that they do not fit neatly into the data distribution.
The presence of outliers can cause problems. For example, a single variable may have an outlier far from the mass of examples, which can skew summary statistics such as the mean and variance.
Fitting a machine learning model may require the identification and removal of outliers as a data preparation technique.
The process of identifying outliers in a dataset is generally referred to as anomaly detection, where the outliers are “anomalies,” and the rest of the data is “normal.” Outlier detection or anomaly detection is a challenging problem and is comprised of a range of techniques.
In machine learning, one approach to tackling the problem of anomaly detection is one-class classification.
One-Class Classification, or OCC for short, involves fitting a model on the “normal” data and predicting whether new data is normal or an outlier/anomaly.
A one-class classifier aims at capturing characteristics of training instances, in order to be able to distinguish between them and potential outliers to appear.
— Page 139, Learning from Imbalanced Data Sets, 2018.
A one-class classifier is fit on a training dataset that only has examples from the normal class. Once prepared, the model is used to classify new examples as either normal or not-normal, i.e. outliers or anomalies.
One-class classification techniques can be used for binary (two-class) imbalanced classification problems where the negative case (class 0) is taken as “normal” and the positive case (class 1) is taken as an outlier or anomaly.
- Negative Case: Normal or inlier.
- Positive Case: Anomaly or outlier.
Given the nature of the approach, one-class classifications are most suited for those tasks where the positive cases don’t have a consistent pattern or structure in the feature space, making it hard for other classification algorithms to learn a class boundary. Instead, treating the positive cases as outliers, it allows one-class classifiers to ignore the task of discrimination and instead focus on deviations from normal or what is expected.
This solution has proven to be especially useful when the minority class lack any structure, being predominantly composed of small disjuncts or noisy instances.
— Page 139, Learning from Imbalanced Data Sets, 2018.
It may also be appropriate where the number of positive cases in the training set is so few that they are not worth including in the model, such as a few tens of examples or fewer. Or for problems where no examples of positive cases can be collected prior to training a model.
To be clear, this adaptation of one-class classification algorithms for imbalanced classification is unusual but can be effective on some problems. The downside of this approach is that any examples of outliers (positive cases) we have during training are not used by the one-class classifier and are discarded. This suggests that perhaps an inverse modeling of the problem (e.g. model the positive case as normal) could be tried in parallel. It also suggests that the one-class classifier could provide an input to an ensemble of algorithms, each of which uses the training dataset in different ways.
One must remember that the advantages of one-class classifiers come at a price of discarding all of available information about the majority class. Therefore, this solution should be used carefully and may not fit some specific applications.
— Page 140, Learning from Imbalanced Data Sets, 2018.
The scikit-learn library provides a handful of common one-class classification algorithms intended for use in outlier or anomaly detection and change detection, such as One-Class SVM, Isolation Forest, Elliptic Envelope, and Local Outlier Factor.
In the following sections, we will take a look at each in turn.
Before we do, we will devise a binary classification dataset to demonstrate the algorithms. We will use the make_classification() scikit-learn function to create 10,000 examples with 10 examples in the minority class and 9,990 in the majority class, or a 0.1 percent vs. 99.9 percent, or about 1:1000 class distribution.
The example below creates and summarizes this dataset.
# Generate and plot a synthetic imbalanced classification dataset from collections import Counter from sklearn.datasets import make_classification from matplotlib import pyplot from numpy import where # define dataset X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0, n_clusters_per_class=1, weights=[0.999], flip_y=0, random_state=4) # summarize class distribution counter = Counter(y) print(counter) # scatter plot of examples by class label for label, _ in counter.items(): row_ix = where(y == label)[0] pyplot.scatter(X[row_ix, 0], X[row_ix, 1], label=str(label)) pyplot.legend() pyplot.show()
Running the example first summarizes the class distribution, confirming the imbalance was created as expected.
Counter({0: 9990, 1: 10})
Next, a scatter plot is created and examples are plotted as points colored by their class label, showing a large mass for the majority class (blue) and a few dots for the minority class (orange).
This severe class imbalance with so few examples in the positive class and the unstructured nature of the few examples in the positive class might make a good basis for using one-class classification methods.
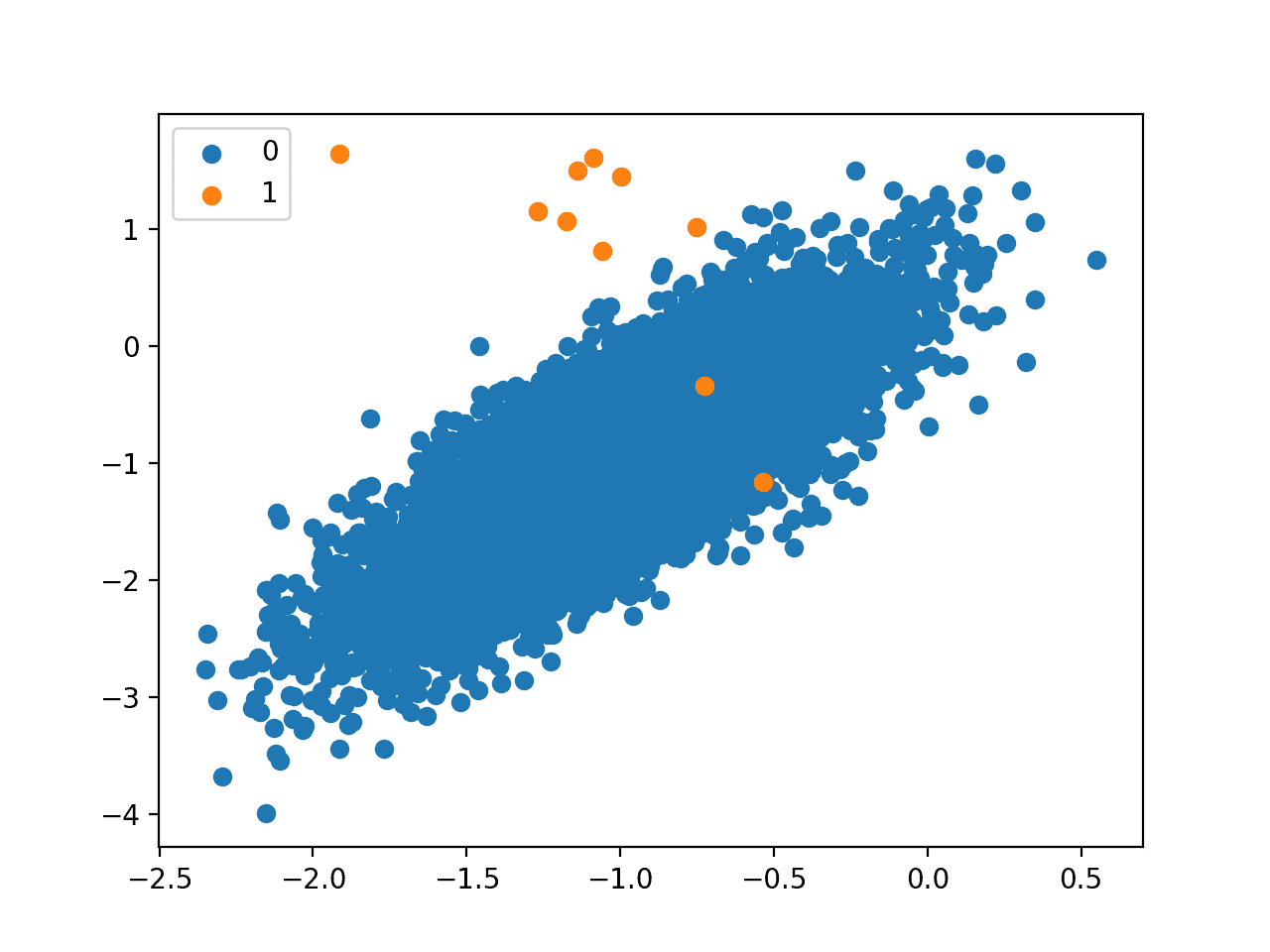
Scatter Plot of a Binary Classification Problem With a 1 to 1000 Class Imbalance
Want to Get Started With Imbalance Classification?
Take my free 7-day email crash course now (with sample code).
Click to sign-up and also get a free PDF Ebook version of the course.
One-Class Support Vector Machines
The support vector machine, or SVM, algorithm developed initially for binary classification can be used for one-class classification.
If used for imbalanced classification, it is a good idea to evaluate the standard SVM and weighted SVM on your dataset before testing the one-class version.
When modeling one class, the algorithm captures the density of the majority class and classifies examples on the extremes of the density function as outliers. This modification of SVM is referred to as One-Class SVM.
… an algorithm that computes a binary function that is supposed to capture regions in input space where the probability density lives (its support), that is, a function such that most of the data will live in the region where the function is nonzero.
— Estimating the Support of a High-Dimensional Distribution, 2001.
The scikit-learn library provides an implementation of one-class SVM in the OneClassSVM class.
The main difference from a standard SVM is that it is fit in an unsupervised manner and does not provide the normal hyperparameters for tuning the margin like C. Instead, it provides a hyperparameter “nu” that controls the sensitivity of the support vectors and should be tuned to the approximate ratio of outliers in the data, e.g. 0.01%.
... # define outlier detection model model = OneClassSVM(gamma='scale', nu=0.01)
The model can be fit on all examples in the training dataset or just those examples in the majority class. Perhaps try both on your problem.
In this case, we will try fitting on just those examples in the training set that belong to the majority class.
# fit on majority class trainX = trainX[trainy==0] model.fit(trainX)
Once fit, the model can be used to identify outliers in new data.
When calling the predict() function on the model, it will output a +1 for normal examples, so-called inliers, and a -1 for outliers.
- Inlier Prediction: +1
- Outlier Prediction: -1
... # detect outliers in the test set yhat = model.predict(testX)
If we want to evaluate the performance of the model as a binary classifier, we must change the labels in the test dataset from 0 and 1 for the majority and minority classes respectively, to +1 and -1.
... # mark inliers 1, outliers -1 testy[testy == 1] = -1 testy[testy == 0] = 1
We can then compare the predictions from the model to the expected target values and calculate a score. Given that we have crisp class labels, we might use a score like precision, recall, or a combination of both, such as the F-measure (F1-score).
In this case, we will use F-measure score, which is the harmonic mean of precision and recall. We can calculate the F-measure using the f1_score() function and specify the label of the minority class as -1 via the “pos_label” argument.
...
# calculate score
score = f1_score(testy, yhat, pos_label=-1)
print('F1 Score: %.3f' % score)
Tying this together, we can evaluate the one-class SVM algorithm on our synthetic dataset. We will split the dataset in two and use half to train the model in an unsupervised manner and the other half to evaluate it.
The complete example is listed below.
# one-class svm for imbalanced binary classification
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.svm import OneClassSVM
# generate dataset
X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0,
n_clusters_per_class=1, weights=[0.999], flip_y=0, random_state=4)
# split into train/test sets
trainX, testX, trainy, testy = train_test_split(X, y, test_size=0.5, random_state=2, stratify=y)
# define outlier detection model
model = OneClassSVM(gamma='scale', nu=0.01)
# fit on majority class
trainX = trainX[trainy==0]
model.fit(trainX)
# detect outliers in the test set
yhat = model.predict(testX)
# mark inliers 1, outliers -1
testy[testy == 1] = -1
testy[testy == 0] = 1
# calculate score
score = f1_score(testy, yhat, pos_label=-1)
print('F1 Score: %.3f' % score)
Running the example fits the model on the input examples from the majority class in the training set. The model is then used to classify examples in the test set as inliers and outliers.
Your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a number of times.
In this case, an F1 score of 0.123 is achieved.
F1 Score: 0.123
Isolation Forest
Isolation Forest, or iForest for short, is a tree-based anomaly detection algorithm.
… Isolation Forest (iForest) which detects anomalies purely based on the concept of isolation without employing any distance or density measure
— Isolation-Based Anomaly Detection, 2012.
It is based on modeling the normal data in such a way to isolate anomalies that are both few in number and different in the feature space.
… our proposed method takes advantage of two anomalies’ quantitative properties: i) they are the minority consisting of fewer instances and ii) they have attribute-values that are very different from those of normal instances.
— Isolation Forest, 2008.
Tree structures are created to isolate anomalies. The result is that isolated examples have a relatively short depth in the trees, whereas normal data is less isolated and has a greater depth in the trees.
… a tree structure can be constructed effectively to isolate every single instance. Because of their susceptibility to isolation, anomalies are isolated closer to the root of the tree; whereas normal points are isolated at the deeper end of the tree.
— Isolation Forest, 2008.
The scikit-learn library provides an implementation of Isolation Forest in the IsolationForest class.
Perhaps the most important hyperparameters of the model are the “n_estimators” argument that sets the number of trees to create and the “contamination” argument, which is used to help define the number of outliers in the dataset.
We know the contamination is about 0.01 percent positive cases to negative cases, so we can set the “contamination” argument to be 0.01.
... # define outlier detection model model = IsolationForest(contamination=0.01, behaviour='new')
The model is probably best trained on examples that exclude outliers. In this case, we fit the model on the input features for examples from the majority class only.
... # fit on majority class trainX = trainX[trainy==0] model.fit(trainX)
Like one-class SVM, the model will predict an inlier with a label of +1 and an outlier with a label of -1, therefore, the labels of the test set must be changed before evaluating the predictions.
Tying this together, the complete example is listed below.
# isolation forest for imbalanced classification
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.ensemble import IsolationForest
# generate dataset
X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0,
n_clusters_per_class=1, weights=[0.999], flip_y=0, random_state=4)
# split into train/test sets
trainX, testX, trainy, testy = train_test_split(X, y, test_size=0.5, random_state=2, stratify=y)
# define outlier detection model
model = IsolationForest(contamination=0.01, behaviour='new')
# fit on majority class
trainX = trainX[trainy==0]
model.fit(trainX)
# detect outliers in the test set
yhat = model.predict(testX)
# mark inliers 1, outliers -1
testy[testy == 1] = -1
testy[testy == 0] = 1
# calculate score
score = f1_score(testy, yhat, pos_label=-1)
print('F1 Score: %.3f' % score)
Running the example fits the isolation forest model on the training dataset in an unsupervised manner, then classifies examples in the test set as inliers and outliers and scores the result.
Your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a number of times.
In this case, an F1 score of 0.154 is achieved.
F1 Score: 0.154
Note: the contamination is quite low and may result in many runs with an F1 Score of 0.0.
To improve the stability of the method on this dataset, try increasing the contamination to 0.05 or even 0.1 and re-run the example.
Minimum Covariance Determinant
If the input variables have a Gaussian distribution, then simple statistical methods can be used to detect outliers.
For example, if the dataset has two input variables and both are Gaussian, then the feature space forms a multi-dimensional Gaussian and knowledge of this distribution can be used to identify values far from the distribution.
This approach can be generalized by defining a hypersphere (ellipsoid) that covers the normal data, and data that falls outside this shape is considered an outlier. An efficient implementation of this technique for multivariate data is known as the Minimum Covariance Determinant, or MCD for short.
It is unusual to have such well-behaved data, but if this is the case for your dataset, or you can use power transforms to make the variables Gaussian, then this approach might be appropriate.
The Minimum Covariance Determinant (MCD) method is a highly robust estimator of multivariate location and scatter, for which a fast algorithm is available. […] It also serves as a convenient and efficient tool for outlier detection.
— Minimum Covariance Determinant and Extensions, 2017.
The scikit-learn library provides access to this method via the EllipticEnvelope class.
It provides the “contamination” argument that defines the expected ratio of outliers to be observed in practice. We know that this is 0.01 percent in our synthetic dataset, so we can set it accordingly.
... # define outlier detection model model = EllipticEnvelope(contamination=0.01)
The model can be fit on the input data from the majority class only in order to estimate the distribution of “normal” data in an unsupervised manner.
... # fit on majority class trainX = trainX[trainy==0] model.fit(trainX)
The model will then be used to classify new examples as either normal (+1) or outliers (-1).
... # detect outliers in the test set yhat = model.predict(testX)
Tying this together, the complete example of using the elliptic envelope outlier detection model for imbalanced classification on our synthetic binary classification dataset is listed below.
# elliptic envelope for imbalanced classification
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.covariance import EllipticEnvelope
# generate dataset
X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0,
n_clusters_per_class=1, weights=[0.999], flip_y=0, random_state=4)
# split into train/test sets
trainX, testX, trainy, testy = train_test_split(X, y, test_size=0.5, random_state=2, stratify=y)
# define outlier detection model
model = EllipticEnvelope(contamination=0.01)
# fit on majority class
trainX = trainX[trainy==0]
model.fit(trainX)
# detect outliers in the test set
yhat = model.predict(testX)
# mark inliers 1, outliers -1
testy[testy == 1] = -1
testy[testy == 0] = 1
# calculate score
score = f1_score(testy, yhat, pos_label=-1)
print('F1 Score: %.3f' % score)
Running the example fits the elliptic envelope model on the training dataset in an unsupervised manner, then classifies examples in the test set as inliers and outliers and scores the result.
Your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a number of times.
In this case, an F1 score of 0.157 is achieved.
F1 Score: 0.157
Local Outlier Factor
A simple approach to identifying outliers is to locate those examples that are far from the other examples in the feature space.
This can work well for feature spaces with low dimensionality (few features), although it can become less reliable as the number of features is increased, referred to as the curse of dimensionality.
The local outlier factor, or LOF for short, is a technique that attempts to harness the idea of nearest neighbors for outlier detection. Each example is assigned a scoring of how isolated or how likely it is to be outliers based on the size of its local neighborhood. Those examples with the largest score are more likely to be outliers.
We introduce a local outlier (LOF) for each object in the dataset, indicating its degree of outlier-ness.
— LOF: Identifying Density-based Local Outliers, 2000.
The scikit-learn library provides an implementation of this approach in the LocalOutlierFactor class.
The model can be defined and requires that the expected percentage of outliers in the dataset be indicated, such as 0.01 percent in the case of our synthetic dataset.
... # define outlier detection model model = LocalOutlierFactor(contamination=0.01)
The model is not fit. Instead, a “normal” dataset is used as the basis for identifying outliers in new data via a call to fit_predict().
To use this model to identify outliers in our test dataset, we must first prepare the training dataset to only have input examples from the majority class.
... # get examples for just the majority class trainX = trainX[trainy==0]
Next, we can concatenate these examples with the input examples from the test dataset.
... # create one large dataset composite = vstack((trainX, testX))
We can then make a prediction by calling fit_predict() and retrieve only those labels for the examples in the test set.
... # make prediction on composite dataset yhat = model.fit_predict(composite) # get just the predictions on the test set yhat yhat[len(trainX):]
To make things easier, we can wrap this up into a new function with the name lof_predict() listed below.
# make a prediction with a lof model def lof_predict(model, trainX, testX): # create one large dataset composite = vstack((trainX, testX)) # make prediction on composite dataset yhat = model.fit_predict(composite) # return just the predictions on the test set return yhat[len(trainX):]
The predicted labels will be +1 for normal and -1 for outliers, like the other outlier detection algorithms in scikit-learn.
Tying this together, the complete example of using the LOF outlier detection algorithm for classification with a skewed class distribution is listed below.
# local outlier factor for imbalanced classification
from numpy import vstack
from sklearn.datasets import make_classification
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.neighbors import LocalOutlierFactor
# make a prediction with a lof model
def lof_predict(model, trainX, testX):
# create one large dataset
composite = vstack((trainX, testX))
# make prediction on composite dataset
yhat = model.fit_predict(composite)
# return just the predictions on the test set
return yhat[len(trainX):]
# generate dataset
X, y = make_classification(n_samples=10000, n_features=2, n_redundant=0,
n_clusters_per_class=1, weights=[0.999], flip_y=0, random_state=4)
# split into train/test sets
trainX, testX, trainy, testy = train_test_split(X, y, test_size=0.5, random_state=2, stratify=y)
# define outlier detection model
model = LocalOutlierFactor(contamination=0.01)
# get examples for just the majority class
trainX = trainX[trainy==0]
# detect outliers in the test set
yhat = lof_predict(model, trainX, testX)
# mark inliers 1, outliers -1
testy[testy == 1] = -1
testy[testy == 0] = 1
# calculate score
score = f1_score(testy, yhat, pos_label=-1)
print('F1 Score: %.3f' % score)
Running the example uses the local outlier factor model with the training dataset in an unsupervised manner to classify examples in the test set as inliers and outliers, then scores the result.
Your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a number of times.
In this case, an F1 score of 0.138 is achieved.
F1 Score: 0.138
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Papers
- Estimating the Support of a High-Dimensional Distribution, 2001.
- Isolation Forest, 2008.
- Isolation-Based Anomaly Detection, 2012.
- A Fast Algorithm for the Minimum Covariance Determinant Estimator, 2012.
- Minimum Covariance Determinant and Extensions, 2017.
- LOF: Identifying Density-based Local Outliers, 2000.
Books
- Learning from Imbalanced Data Sets, 2018.
- Imbalanced Learning: Foundations, Algorithms, and Applications, 2013.
APIs
- Novelty and Outlier Detection, scikit-learn API.
- sklearn.svm.OneClassSVM API.
- sklearn.ensemble.IsolationForest API.
- sklearn.covariance.EllipticEnvelope API.
- sklearn.neighbors.LocalOutlierFactor API.
Articles
Summary
In this tutorial, you discovered how to use one-class classification algorithms for datasets with severely skewed class distributions.
Specifically, you learned:
- One-class classification is a field of machine learning that provides techniques for outlier and anomaly detection.
- How to adapt one-class classification algorithms for imbalanced classification with a severely skewed class distribution.
- How to fit and evaluate one-class classification algorithms such as SVM, isolation forest, elliptic envelope and local outlier factor.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post One-Class Classification Algorithms for Imbalanced Datasets appeared first on Machine Learning Mastery.
