Author: Jason Brownlee
Many machine learning algorithms perform better when numerical input variables are scaled to a standard range.
This includes algorithms that use a weighted sum of the input, like linear regression, and algorithms that use distance measures, like k-nearest neighbors.
Standardizing is a popular scaling technique that subtracts the mean from values and divides by the standard deviation, transforming the probability distribution for an input variable to a standard Gaussian (zero mean and unit variance). Standardization can become skewed or biased if the input variable contains outlier values.
To overcome this, the median and interquartile range can be used when standardizing numerical input variables, generally referred to as robust scaling.
In this tutorial, you will discover how to use robust scaler transforms to standardize numerical input variables for classification and regression.
After completing this tutorial, you will know:
- Many machine learning algorithms prefer or perform better when numerical input variables are scaled.
- Robust scaling techniques that use percentiles can be used to scale numerical input variables that contain outliers.
- How to use the RobustScaler to scale numerical input variables using the median and interquartile range.
Let’s get started.

How to Use Robust Scaler Transforms for Machine Learning
Photo by Ray in Manila, some rights reserved.
Tutorial Overview
This tutorial is divided into five parts; they are:
- Scaling Data
- Robust Scaler Transforms
- Sonar Dataset
- IQR Robust Scaler Transform
- Explore Robust Scaler Range
Robust Scaling Data
It is common to scale data prior to fitting a machine learning model.
This is because data often consists of many different input variables or features (columns) and each may have a different range of values or units of measure, such as feet, miles, kilograms, dollars, etc.
If there are input variables that have very large values relative to the other input variables, these large values can dominate or skew some machine learning algorithms. The result is that the algorithms pay most of their attention to the large values and ignore the variables with smaller values.
This includes algorithms that use a weighted sum of inputs like linear regression, logistic regression, and artificial neural networks, as well as algorithms that use distance measures between examples, such as k-nearest neighbors and support vector machines.
As such, it is normal to scale input variables to a common range as a data preparation technique prior to fitting a model.
One approach to data scaling involves calculating the mean and standard deviation of each variable and using these values to scale the values to have a mean of zero and a standard deviation of one, a so-called “standard normal” probability distribution. This process is called standardization and is most useful when input variables have a Gaussian probability distribution.
Standardization is calculated by subtracting the mean value and dividing by the standard deviation.
- value = (value – mean) / stdev
Sometimes an input variable may have outlier values. These are values on the edge of the distribution that may have a low probability of occurrence, yet are overrepresented for some reason. Outliers can skew a probability distribution and make data scaling using standardization difficult as the calculated mean and standard deviation will be skewed by the presence of the outliers.
One approach to standardizing input variables in the presence of outliers is to ignore the outliers from the calculation of the mean and standard deviation, then use the calculated values to scale the variable.
This is called robust standardization or robust data scaling.
This can be achieved by calculating the median (50th percentile) and the 25th and 75th percentiles. The values of each variable then have their median subtracted and are divided by the interquartile range (IQR) which is the difference between the 75th and 25th percentiles.
- value = (value – median) / (p75 – p25)
The resulting variable has a zero mean and median and a standard deviation of 1, although not skewed by outliers and the outliers are still present with the same relative relationships to other values.
Robust Scaler Transforms
The robust scaler transform is available in the scikit-learn Python machine learning library via the RobustScaler class.
The “with_centering” argument controls whether the value is centered to zero (median is subtracted) and defaults to True.
The “with_scaling” argument controls whether the value is scaled to the IQR (standard deviation set to one) or not and defaults to True.
Interestingly, the definition of the scaling range can be specified via the “quantile_range” argument. It takes a tuple of two integers between 0 and 100 and defaults to the percentile values of the IQR, specifically (25, 75). Changing this will change the definition of outliers and the scope of the scaling.
We will take a closer look at how to use the robust scaler transforms on a real dataset.
First, let’s introduce a real dataset.
Sonar Dataset
The sonar dataset is a standard machine learning dataset for binary classification.
It involves 60 real-valued inputs and a two-class target variable. There are 208 examples in the dataset and the classes are reasonably balanced.
A baseline classification algorithm can achieve a classification accuracy of about 53.4 percent using repeated stratified 10-fold cross-validation. Top performance on this dataset is about 88 percent using repeated stratified 10-fold cross-validation.
The dataset describes radar returns of rocks or simulated mines.
You can learn more about the dataset from here:
No need to download the dataset; we will download it automatically from our worked examples.
First, let’s load and summarize the dataset. The complete example is listed below.
# load and summarize the sonar dataset from pandas import read_csv from pandas.plotting import scatter_matrix from matplotlib import pyplot # Load dataset url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv" dataset = read_csv(url, header=None) # summarize the shape of the dataset print(dataset.shape) # summarize each variable print(dataset.describe()) # histograms of the variables dataset.hist() pyplot.show()
Running the example first summarizes the shape of the loaded dataset.
This confirms the 60 input variables, one output variable, and 208 rows of data.
A statistical summary of the input variables is provided showing that values are numeric and range approximately from 0 to 1.
(208, 61)
0 1 2 ... 57 58 59
count 208.000000 208.000000 208.000000 ... 208.000000 208.000000 208.000000
mean 0.029164 0.038437 0.043832 ... 0.007949 0.007941 0.006507
std 0.022991 0.032960 0.038428 ... 0.006470 0.006181 0.005031
min 0.001500 0.000600 0.001500 ... 0.000300 0.000100 0.000600
25% 0.013350 0.016450 0.018950 ... 0.003600 0.003675 0.003100
50% 0.022800 0.030800 0.034300 ... 0.005800 0.006400 0.005300
75% 0.035550 0.047950 0.057950 ... 0.010350 0.010325 0.008525
max 0.137100 0.233900 0.305900 ... 0.044000 0.036400 0.043900
[8 rows x 60 columns]
Finally, a histogram is created for each input variable.
If we ignore the clutter of the plots and focus on the histograms themselves, we can see that many variables have a skewed distribution.
The dataset provides a good candidate for using a robust scaler transform to standardize the data in the presence of skewed distributions and outliers.
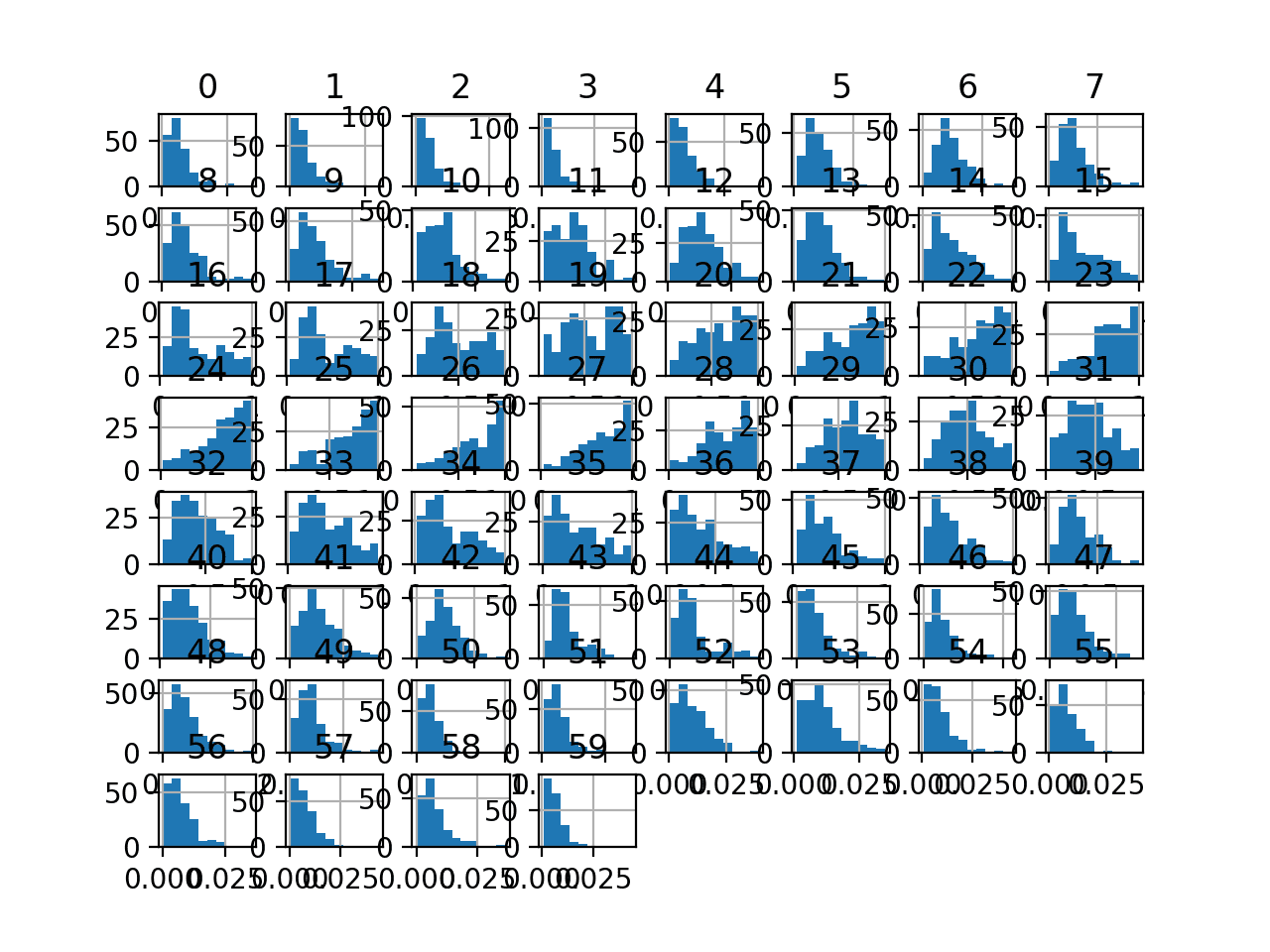
Histogram Plots of Input Variables for the Sonar Binary Classification Dataset
Next, let’s fit and evaluate a machine learning model on the raw dataset.
We will use a k-nearest neighbor algorithm with default hyperparameters and evaluate it using repeated stratified k-fold cross-validation. The complete example is listed below.
# evaluate knn on the raw sonar dataset
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import LabelEncoder
from matplotlib import pyplot
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
# define and configure the model
model = KNeighborsClassifier()
# evaluate the model
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report model performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example evaluates a KNN model on the raw sonar dataset.
We can see that the model achieved a mean classification accuracy of about 79.7 percent, showing that it has skill (better than 53.4 percent) and is in the ball-park of good performance (88 percent).
Accuracy: 0.797 (0.073)
Next, let’s explore a robust scaling transform of the dataset.
IQR Robust Scaler Transform
We can apply the robust scaler to the Sonar dataset directly.
We will use the default configuration and scale values to the IQR. First, a RobustScaler instance is defined with default hyperparameters. Once defined, we can call the fit_transform() function and pass it to our dataset to create a quantile transformed version of our dataset.
... # perform a robust scaler transform of the dataset trans = RobustScaler() data = trans.fit_transform(data)
Let’s try it on our sonar dataset.
The complete example of creating a robust scaler transform of the sonar dataset and plotting histograms of the result is listed below.
# visualize a robust scaler transform of the sonar dataset from pandas import read_csv from pandas import DataFrame from pandas.plotting import scatter_matrix from sklearn.preprocessing import RobustScaler from matplotlib import pyplot # load dataset url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv" dataset = read_csv(url, header=None) # retrieve just the numeric input values data = dataset.values[:, :-1] # perform a robust scaler transform of the dataset trans = RobustScaler() data = trans.fit_transform(data) # convert the array back to a dataframe dataset = DataFrame(data) # summarize print(dataset.describe()) # histograms of the variables dataset.hist() pyplot.show()
Running the example first reports a summary of each input variable.
We can see that the distributions have been adjusted. The median values are now zero and the standard deviation values are now close to 1.0.
0 1 ... 58 59 count 208.000000 208.000000 ... 2.080000e+02 208.000000 mean 0.286664 0.242430 ... 2.317814e-01 0.222527 std 1.035627 1.046347 ... 9.295312e-01 0.927381 min -0.959459 -0.958730 ... -9.473684e-01 -0.866359 25% -0.425676 -0.455556 ... -4.097744e-01 -0.405530 50% 0.000000 0.000000 ... 6.591949e-17 0.000000 75% 0.574324 0.544444 ... 5.902256e-01 0.594470 max 5.148649 6.447619 ... 4.511278e+00 7.115207 [8 rows x 60 columns]
Histogram plots of the variables are created, although the distributions don’t look much different from their original distributions seen in the previous section.
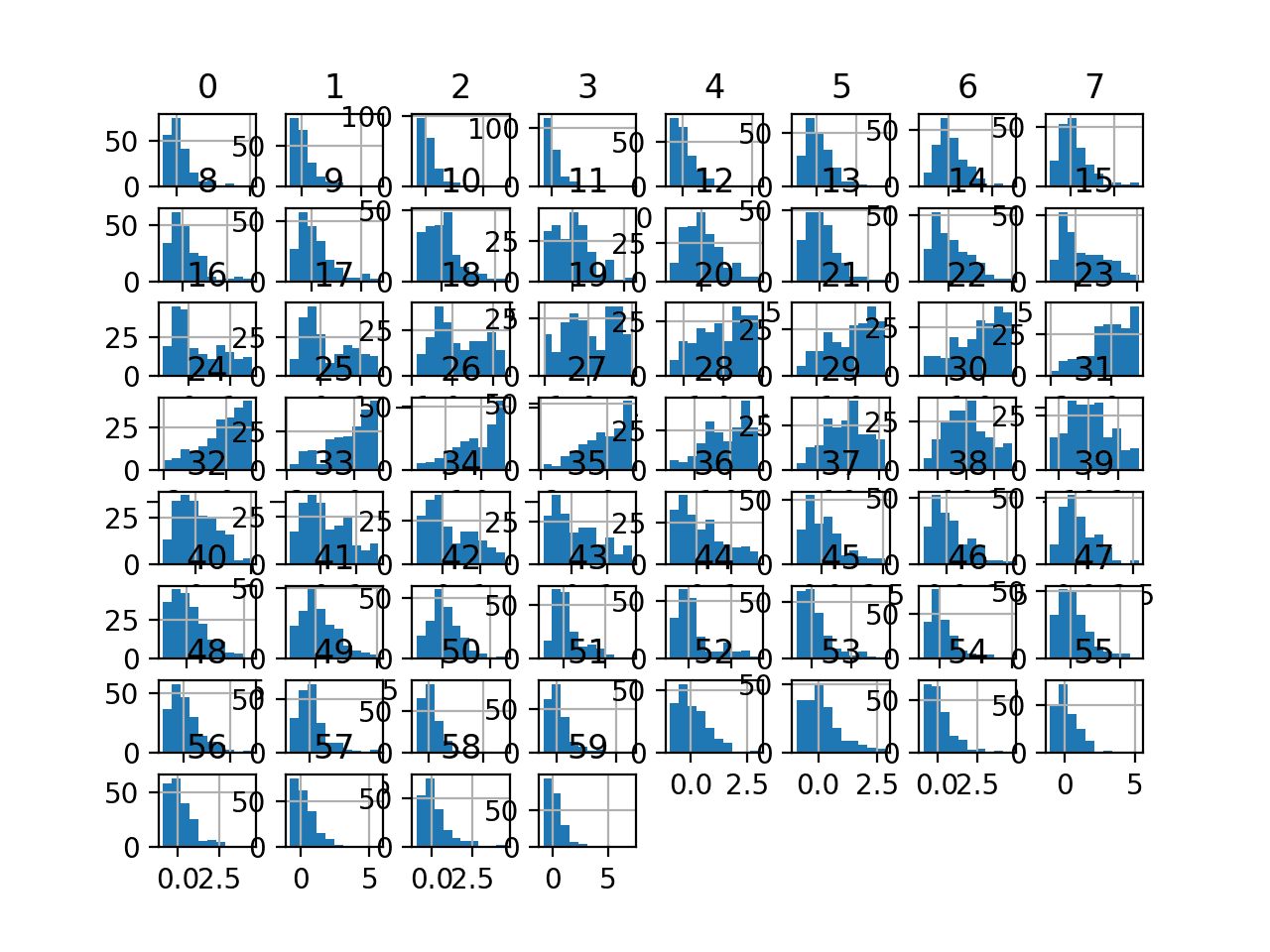
Histogram Plots of Robust Scaler Transformed Input Variables for the Sonar Dataset
Next, let’s evaluate the same KNN model as the previous section, but in this case on a robust scaler transform of the dataset.
The complete example is listed below.
# evaluate knn on the sonar dataset with robust scaler transform
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import RobustScaler
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
# define the pipeline
trans = RobustScaler(with_centering=False, with_scaling=True)
model = KNeighborsClassifier()
pipeline = Pipeline(steps=[('t', trans), ('m', model)])
# evaluate the pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(pipeline, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report pipeline performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example, we can see that the robust scaler transform results in a lift in performance from 79.7 percent accuracy without the transform to about 81.9 percent with the transform.
Accuracy: 0.819 (0.076)
Next, let’s explore the effect of different scaling ranges.
Explore Robust Scaler Range
The range used to scale each variable is chosen by default as the IQR is bounded by the 25th and 75th percentiles.
This is specified by the “quantile_range” argument as a tuple.
Other values can be specified and might improve the performance of the model, such as a wider range, allowing fewer values to be considered outliers, or a more narrow range, allowing more values to be considered outliers.
The example below explores the effect of different definitions of the range from 1st to the 99th percentiles to 30th to 70th percentiles.
The complete example is listed below.
# explore the scaling range of the robust scaler transform
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import RobustScaler
from sklearn.preprocessing import LabelEncoder
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# get the dataset
def get_dataset():
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
return X, y
# get a list of models to evaluate
def get_models():
models = dict()
for value in [1, 5, 10, 15, 20, 25, 30]:
# define the pipeline
trans = RobustScaler(quantile_range=(value, 100-value))
model = KNeighborsClassifier()
models[str(value)] = Pipeline(steps=[('t', trans), ('m', model)])
return models
# evaluate a give model using cross-validation
def evaluate_model(model):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
return scores
# define dataset
X, y = get_dataset()
# get the models to evaluate
models = get_models()
# evaluate the models and store results
results, names = list(), list()
for name, model in models.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print('>%s %.3f (%.3f)' % (name, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=names, showmeans=True)
pyplot.show()
Running the example reports the mean classification accuracy for each value-defined IQR range.
We can see that the default of 25th to 75th percentile achieves the best results, although the values of 20-80 and 30-70 achieve results that are very similar.
>1 0.818 (0.069) >5 0.813 (0.085) >10 0.812 (0.076) >15 0.811 (0.081) >20 0.811 (0.080) >25 0.819 (0.076) >30 0.816 (0.072)
Box and whisker plots are created to summarize the classification accuracy scores for each IQR range.
We can see a marked difference in the distribution and mean accuracy with the larger ranges of 25-75 and 30-70 percentiles.
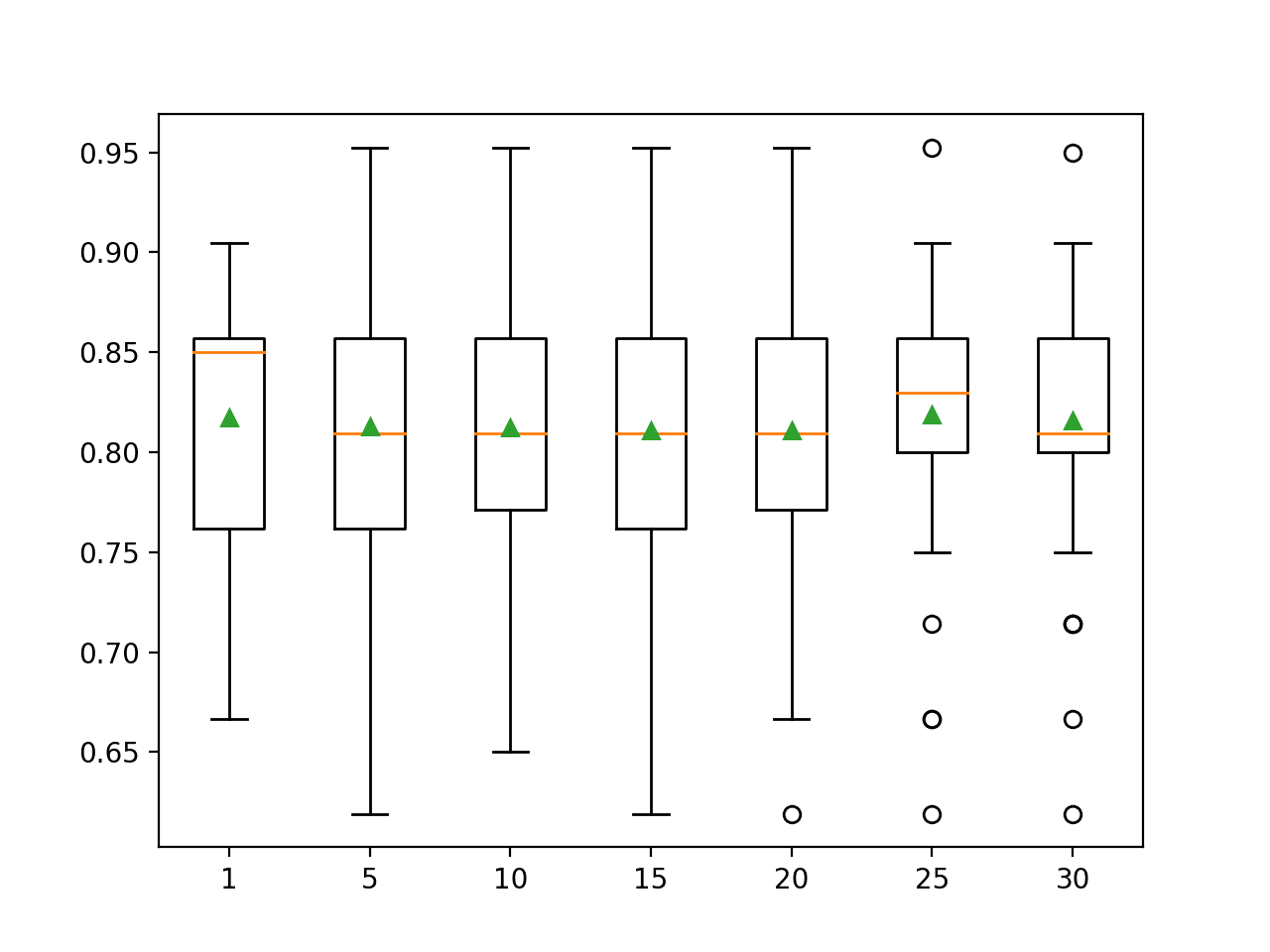
Box Plots of Robust Scaler IQR Range vs Classification Accuracy of KNN on the Sonar Dataset
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Tutorials
APIs
- Standardization, or mean removal and variance scaling, scikit-learn.
- sklearn.preprocessing.RobustScaler API.
Articles
Summary
In this tutorial, you discovered how to use robust scaler transforms to standardize numerical input variables for classification and regression.
Specifically, you learned:
- Many machine learning algorithms prefer or perform better when numerical input variables are scaled.
- Robust scaling techniques that use percentiles can be used to scale numerical input variables that contain outliers.
- How to use the RobustScaler to scale numerical input variables using the median and interquartile range.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post How to Scale Data With Outliers for Machine Learning appeared first on Machine Learning Mastery.
