Author: Jason Brownlee
Feature selection is the process of identifying and selecting a subset of input features that are most relevant to the target variable.
Feature selection is often straightforward when working with real-valued input and output data, such as using the Pearson’s correlation coefficient, but can be challenging when working with numerical input data and a categorical target variable.
The two most commonly used feature selection methods for numerical input data when the target variable is categorical (e.g. classification predictive modeling) are the ANOVA f-test statistic and the mutual information statistic.
In this tutorial, you will discover how to perform feature selection with numerical input data for classification.
After completing this tutorial, you will know:
- The diabetes predictive modeling problem with numerical inputs and binary classification target variables.
- How to evaluate the importance of numerical features using the ANOVA f-test and mutual information statistics.
- How to perform feature selection for numerical data when fitting and evaluating a classification model.
Let’s get started.

How to Perform Feature Selection With Numerical Input Data
Photo by Susanne Nilsson, some rights reserved.
Tutorial Overview
This tutorial is divided into four parts; they are:
- Diabetes Numerical Dataset
- Numerical Feature Selection
- ANOVA f-test Feature Selection
- Mutual Information Feature Selection
- Modeling With Selected Features
- Model Built Using All Features
- Model Built Using ANOVA f-test Features
- Model Built Using Mutual Information Features
- Tune the Number of Selected Features
Diabetes Numerical Dataset
As the basis of this tutorial, we will use the so-called “diabetes” dataset that has been widely studied as a machine learning dataset since 1990.
The dataset classifies patients’ data as either an onset of diabetes within five years or not. There are 768 examples and eight input variables. It is a binary classification problem.
A naive model can achieve an accuracy of about 65 percent on this dataset. A good score is about 77 percent +/- 5 percent. We will aim for this region but note that the models in this tutorial are not optimized; they are designed to demonstrate feature selection schemes.
You can download the dataset and save the file as “pima-indians-diabetes.csv” in your current working directory.
- Diabetes Dataset (pima-indians-diabetes.csv)
- Diabetes Dataset Description (pima-indians-diabetes.names)
Looking at the data, we can see that all nine input variables are numerical.
6,148,72,35,0,33.6,0.627,50,1 1,85,66,29,0,26.6,0.351,31,0 8,183,64,0,0,23.3,0.672,32,1 1,89,66,23,94,28.1,0.167,21,0 0,137,40,35,168,43.1,2.288,33,1 ...
We can load this dataset into memory using the Pandas library.
... # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values
Once loaded, we can split the columns into input (X) and output (y) for modeling.
... # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1]
We can tie all of this together into a helpful function that we can reuse later.
# load the dataset def load_dataset(filename): # load the dataset as a pandas DataFrame data = read_csv(filename, header=None) # retrieve numpy array dataset = data.values # split into input (X) and output (y) variables X = dataset[:, :-1] y = dataset[:,-1] return X, y
Once loaded, we can split the data into training and test sets so we can fit and evaluate a learning model.
We will use the train_test_split() function form scikit-learn and use 67 percent of the data for training and 33 percent for testing.
...
# load the dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
Tying all of these elements together, the complete example of loading, splitting, and summarizing the raw categorical dataset is listed below.
# load and summarize the dataset
from pandas import read_csv
from sklearn.model_selection import train_test_split
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# load the dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# summarize
print('Train', X_train.shape, y_train.shape)
print('Test', X_test.shape, y_test.shape)
Running the example reports the size of the input and output elements of the train and test sets.
We can see that we have 514 examples for training and 254 for testing.
Train (514, 8) (514, 1) Test (254, 8) (254, 1)
Now that we have loaded and prepared the diabetes dataset, we can explore feature selection.
Numerical Feature Selection
There are two popular feature selection techniques that can be used for numerical input data and a categorical (class) target variable.
They are:
- ANOVA-f Statistic.
- Mutual Information Statistics.
Let’s take a closer look at each in turn.
ANOVA f-test Feature Selection
ANOVA is an acronym for “analysis of variance” and is a parametric statistical hypothesis test for determining whether the means from two or more samples of data (often three or more) come from the same distribution or not.
An F-statistic, or F-test, is a class of statistical tests that calculate the ratio between variances values, such as the variance from two different samples or the explained and unexplained variance by a statistical test, like ANOVA. The ANOVA method is a type of F-statistic referred to here as an ANOVA f-test.
Importantly, ANOVA is used when one variable is numeric and one is categorical, such as numerical input variables and a classification target variable in a classification task.
The results of this test can be used for feature selection where those features that are independent of the target variable can be removed from the dataset.
When the outcome is numeric, and […] the predictor has more than two levels, the traditional ANOVA F-statistic can be calculated.
— Page 242, Feature Engineering and Selection, 2019.
The scikit-learn machine library provides an implementation of the ANOVA f-test in the f_classif() function. This function can be used in a feature selection strategy, such as selecting the top k most relevant features (largest values) via the SelectKBest class.
For example, we can define the SelectKBest class to use the f_classif() function and select all features, then transform the train and test sets.
... # configure to select all features fs = SelectKBest(score_func=f_classif, k='all') # learn relationship from training data fs.fit(X_train, y_train) # transform train input data X_train_fs = fs.transform(X_train) # transform test input data X_test_fs = fs.transform(X_test)
We can then print the scores for each variable (larger is better) and plot the scores for each variable as a bar graph to get an idea of how many features we should select.
...
# what are scores for the features
for i in range(len(fs.scores_)):
print('Feature %d: %f' % (i, fs.scores_[i]))
# plot the scores
pyplot.bar([i for i in range(len(fs.scores_))], fs.scores_)
pyplot.show()
Tying this together with the data preparation for the diabetes dataset in the previous section, the complete example is listed below.
# example of anova f-test feature selection for numerical data
from pandas import read_csv
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif
from matplotlib import pyplot
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select all features
fs = SelectKBest(score_func=f_classif, k='all')
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# what are scores for the features
for i in range(len(fs.scores_)):
print('Feature %d: %f' % (i, fs.scores_[i]))
# plot the scores
pyplot.bar([i for i in range(len(fs.scores_))], fs.scores_)
pyplot.show()
Running the example first prints the scores calculated for each input feature and the target variable.
Note that your specific results may differ given the stochastic nature of the learning algorithm. Try running the example a few times.
In this case, we can see that some features stand out as perhaps being more relevant than others, with much larger test statistic values.
Perhaps features 1, 5, and 7 are most relevant.
Feature 0: 16.527385 Feature 1: 131.325562 Feature 2: 0.042371 Feature 3: 1.415216 Feature 4: 12.778966 Feature 5: 49.209523 Feature 6: 13.377142 Feature 7: 25.126440
A bar chart of the feature importance scores for each input feature is created.
This clearly shows that feature 1 might be the most relevant (according to test) and that perhaps six of the eight input features are the more relevant.
We could set k=6 when configuring the SelectKBest to select these top four features.
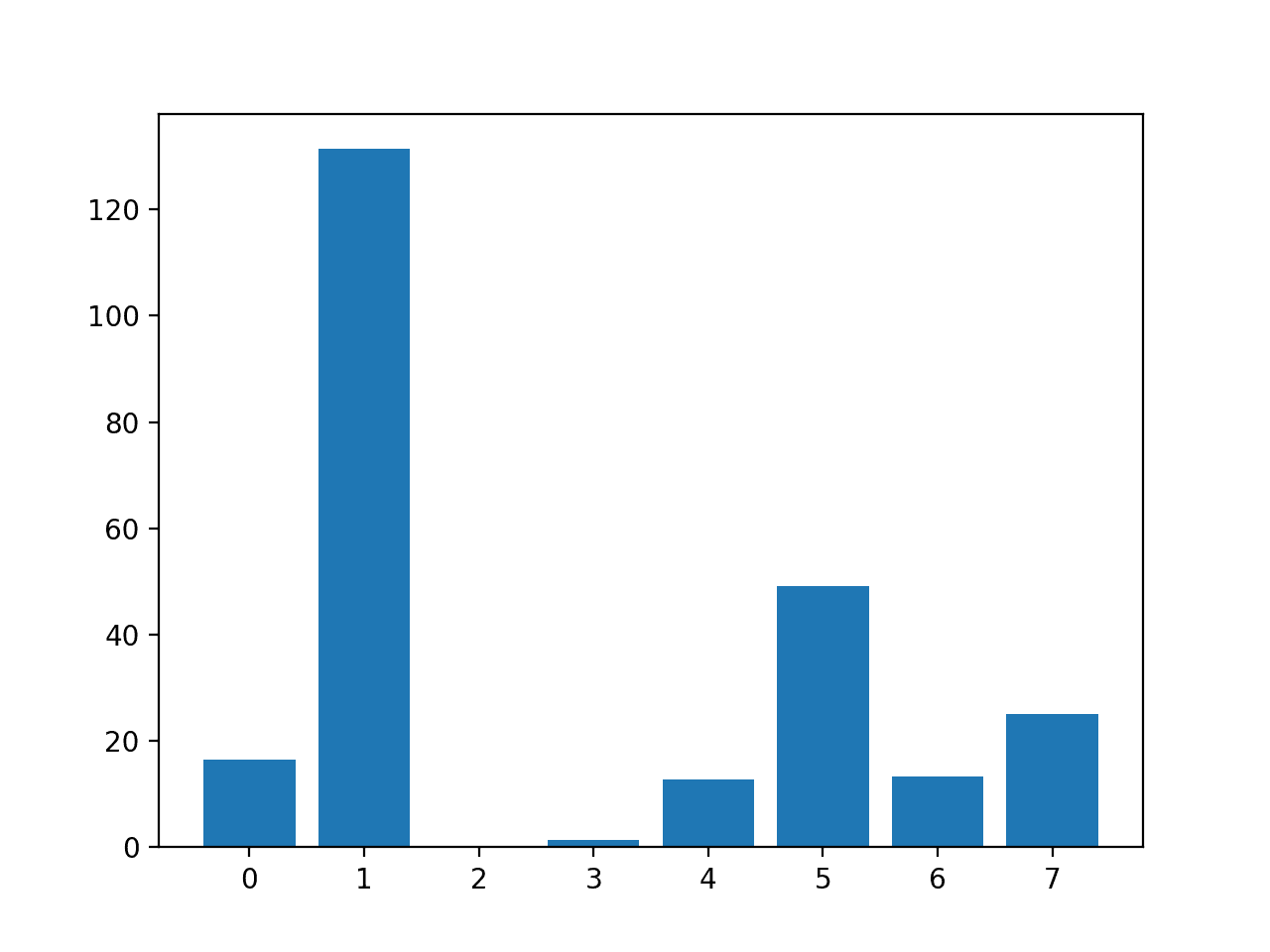
Bar Chart of the Input Features (x) vs The ANOVA f-test Feature Importance (y)
Mutual Information Feature Selection
Mutual information from the field of information theory is the application of information gain (typically used in the construction of decision trees) to feature selection.
Mutual information is calculated between two variables and measures the reduction in uncertainty for one variable given a known value of the other variable.
You can learn more about mutual information in the following tutorial.
Mutual information is straightforward when considering the distribution of two discrete (categorical or ordinal) variables, such as categorical input and categorical output data. Nevertheless, it can be adapted for use with numerical input and categorical output.
For technical details on how this can be achieved, see the 2014 paper titled “Mutual Information between Discrete and Continuous Data Sets.”
The scikit-learn machine learning library provides an implementation of mutual information for feature selection with numeric input and categorical output variables via the mutual_info_classif() function.
Like f_classif(), it can be used in the SelectKBest feature selection strategy (and other strategies).
... # configure to select all features fs = SelectKBest(score_func=mutual_info_classif, k='all') # learn relationship from training data fs.fit(X_train, y_train) # transform train input data X_train_fs = fs.transform(X_train) # transform test input data X_test_fs = fs.transform(X_test)
We can perform feature selection using mutual information on the diabetes dataset and print and plot the scores (larger is better) as we did in the previous section.
The complete example of using mutual information for numerical feature selection is listed below.
# example of mutual information feature selection for numerical input data
from pandas import read_csv
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import mutual_info_classif
from matplotlib import pyplot
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select all features
fs = SelectKBest(score_func=mutual_info_classif, k='all')
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# what are scores for the features
for i in range(len(fs.scores_)):
print('Feature %d: %f' % (i, fs.scores_[i]))
# plot the scores
pyplot.bar([i for i in range(len(fs.scores_))], fs.scores_)
pyplot.show()
Running the example first prints the scores calculated for each input feature and the target variable.
Note: your specific results may differ. Try running the example a few times.
In this case, we can see that some of the features have a modestly low score, suggesting that perhaps they can be removed.
Perhaps features 1 and 5 are most relevant.
Feature 1: 0.118431 Feature 2: 0.019966 Feature 3: 0.041791 Feature 4: 0.019858 Feature 5: 0.084719 Feature 6: 0.018079 Feature 7: 0.033098
A bar chart of the feature importance scores for each input feature is created.
Importantly, a different mixture of features is promoted.
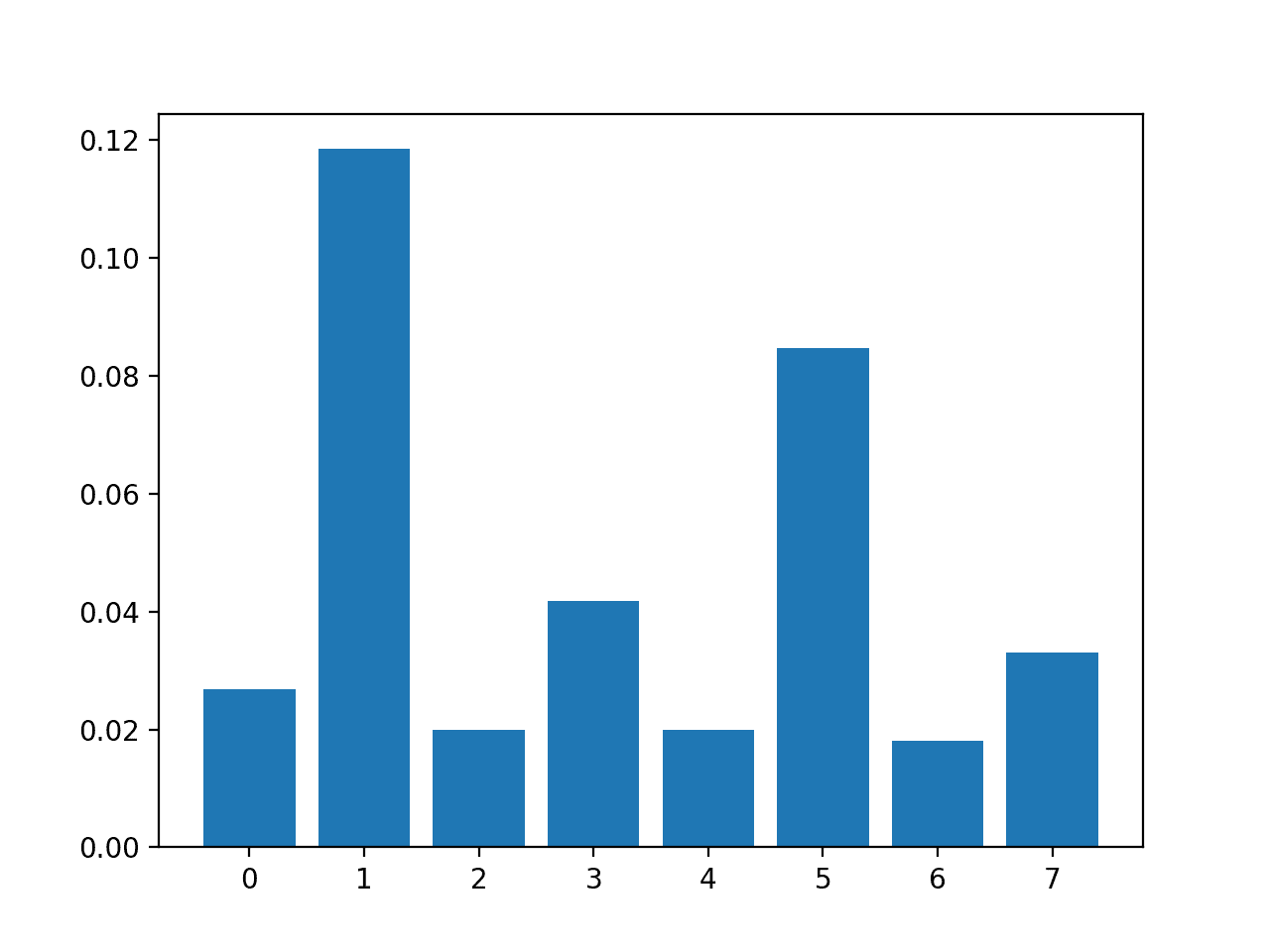
Bar Chart of the Input Features (x) vs. the Mutual Information Feature Importance (y)
Now that we know how to perform feature selection on numerical input data for a classification predictive modeling problem, we can try developing a model using the selected features and compare the results.
Modeling With Selected Features
There are many different techniques for scoring features and selecting features based on scores; how do you know which one to use?
A robust approach is to evaluate models using different feature selection methods (and numbers of features) and select the method that results in a model with the best performance.
In this section, we will evaluate a Logistic Regression model with all features compared to a model built from features selected by ANOVA f-test and those features selected via mutual information.
Logistic regression is a good model for testing feature selection methods as it can perform better if irrelevant features are removed from the model.
Model Built Using All Features
As a first step, we will evaluate a LogisticRegression model using all the available features.
The model is fit on the training dataset and evaluated on the test dataset.
The complete example is listed below.
# evaluation of a model using all input features
from pandas import read_csv
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# load the dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# fit the model
model = LogisticRegression(solver='liblinear')
model.fit(X_train, y_train)
# evaluate the model
yhat = model.predict(X_test)
# evaluate predictions
accuracy = accuracy_score(y_test, yhat)
print('Accuracy: %.2f' % (accuracy*100))
Running the example prints the accuracy of the model on the training dataset.
Note: your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a few times.
In this case, we can see that the model achieves a classification accuracy of about 77 percent.
We would prefer to use a subset of features that achieves a classification accuracy that is as good or better than this.
Accuracy: 77.56
Model Built Using ANOVA f-test Features
We can use the ANOVA f-test to score the features and select the four most relevant features.
The select_features() function below is updated to achieve this.
# feature selection def select_features(X_train, y_train, X_test): # configure to select a subset of features fs = SelectKBest(score_func=f_classif, k=4) # learn relationship from training data fs.fit(X_train, y_train) # transform train input data X_train_fs = fs.transform(X_train) # transform test input data X_test_fs = fs.transform(X_test) return X_train_fs, X_test_fs, fs
The complete example of evaluating a logistic regression model fit and evaluated on data using this feature selection method is listed below.
# evaluation of a model using 4 features chosen with anova f-test
from pandas import read_csv
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select a subset of features
fs = SelectKBest(score_func=f_classif, k=4)
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# fit the model
model = LogisticRegression(solver='liblinear')
model.fit(X_train_fs, y_train)
# evaluate the model
yhat = model.predict(X_test_fs)
# evaluate predictions
accuracy = accuracy_score(y_test, yhat)
print('Accuracy: %.2f' % (accuracy*100))
Running the example reports the performance of the model on just four of the eight input features selected using the ANOVA f-test statistic.
Note: your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a few times.
In this case, we see that the model achieved an accuracy of about 78.74 percent, a lift in performance compared to the baseline that achieved 77.56 percent.
Accuracy: 78.74
Model Built Using Mutual Information Features
We can repeat the experiment and select the top four features using a mutual information statistic.
The updated version of the select_features() function to achieve this is listed below.
# feature selection def select_features(X_train, y_train, X_test): # configure to select a subset of features fs = SelectKBest(score_func=mutual_info_classif, k=4) # learn relationship from training data fs.fit(X_train, y_train) # transform train input data X_train_fs = fs.transform(X_train) # transform test input data X_test_fs = fs.transform(X_test) return X_train_fs, X_test_fs, fs
The complete example of using mutual information for feature selection to fit a logistic regression model is listed below.
# evaluation of a model using 4 features chosen with mutual information
from pandas import read_csv
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import mutual_info_classif
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# feature selection
def select_features(X_train, y_train, X_test):
# configure to select a subset of features
fs = SelectKBest(score_func=mutual_info_classif, k=4)
# learn relationship from training data
fs.fit(X_train, y_train)
# transform train input data
X_train_fs = fs.transform(X_train)
# transform test input data
X_test_fs = fs.transform(X_test)
return X_train_fs, X_test_fs, fs
# load the dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.33, random_state=1)
# feature selection
X_train_fs, X_test_fs, fs = select_features(X_train, y_train, X_test)
# fit the model
model = LogisticRegression(solver='liblinear')
model.fit(X_train_fs, y_train)
# evaluate the model
yhat = model.predict(X_test_fs)
# evaluate predictions
accuracy = accuracy_score(y_test, yhat)
print('Accuracy: %.2f' % (accuracy*100))
Running the example fits the model on the four top selected features chosen using mutual information.
Note that your specific results may vary given the stochastic nature of the learning algorithm. Try running the example a few times.
In this case, we can make no difference compared to the baseline model. This is interesting as we know the method chose a different four features compared to the previous method.
Accuracy: 77.56
Tune the Number of Selected Features
In the previous example, we selected four features, but how do we know that is a good or best number of features to select?
Instead of guessing, we can systematically test a range of different numbers of selected features and discover which results in the best performing model. This is called a grid search, where the k argument to the SelectKBest class can be tuned.
It is good practice to evaluate model configurations on classification tasks using repeated stratified k-fold cross-validation. We will use three repeats of 10-fold cross-validation via the RepeatedStratifiedKFold class.
... # define the evaluation method cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
We can define a Pipeline that correctly prepares the feature selection transform on the training set and applies it to the train set and test set for each fold of the cross-validation.
In this case, we will use the ANOVA f-test statistical method for selecting features.
...
# define the pipeline to evaluate
model = LogisticRegression(solver='liblinear')
fs = SelectKBest(score_func=f_classif)
pipeline = Pipeline(steps=[('anova',fs), ('lr', model)])
We can then define the grid of values to evaluate as 1 to 8.
Note that the grid is a dictionary of parameters to values to search, and given that we are using a Pipeline, we can access the SelectKBest object via the name we gave it, ‘anova‘, and then the parameter name ‘k‘, separated by two underscores, or ‘anova__k‘.
... # define the grid grid = dict() grid['anova__k'] = [i+1 for i in range(X.shape[1])]
We can then define and run the search.
... # define the grid search search = GridSearchCV(pipeline, grid, scoring='accuracy', n_jobs=-1, cv=cv) # perform the search results = search.fit(X, y)
Tying this together, the complete example is listed below.
# compare different numbers of features selected using anova f-test
from pandas import read_csv
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import Pipeline
from sklearn.model_selection import GridSearchCV
from matplotlib import pyplot
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# define dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# define the evaluation method
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
# define the pipeline to evaluate
model = LogisticRegression(solver='liblinear')
fs = SelectKBest(score_func=f_classif)
pipeline = Pipeline(steps=[('anova',fs), ('lr', model)])
# define the grid
grid = dict()
grid['anova__k'] = [i+1 for i in range(X.shape[1])]
# define the grid search
search = GridSearchCV(pipeline, grid, scoring='accuracy', n_jobs=-1, cv=cv)
# perform the search
results = search.fit(X, y)
# summarize best
print('Best Mean Accuracy: %.3f' % results.best_score_)
print('Best Config: %s' % results.best_params_)
Running the example grid searches different numbers of selected features using ANOVA f-test, where each modeling pipeline is evaluated using repeated cross-validation.
Your specific results may vary given the stochastic nature of the learning algorithm and evaluating procedure. Try running the example a few times.
In this case, we can see that the best number of selected features is seven; that achieves an accuracy of about 77 percent.
Best Mean Accuracy: 0.770
Best Config: {'anova__k': 7}
We might want to see the relationship between the number of selected features and classification accuracy. In this relationship, we may expect that more features result in a better performance to a point.
This relationship can be explored by manually evaluating each configuration of k for the SelectKBest from 1 to 8, gathering the sample of accuracy scores, and plotting the results using box and whisker plots side-by-side. The spread and mean of these box plots would be expected to show any interesting relationship between the number of selected features and the classification accuracy of the pipeline.
The complete example of achieving this is listed below.
# compare different numbers of features selected using anova f-test
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# load the dataset
def load_dataset(filename):
# load the dataset as a pandas DataFrame
data = read_csv(filename, header=None)
# retrieve numpy array
dataset = data.values
# split into input (X) and output (y) variables
X = dataset[:, :-1]
y = dataset[:,-1]
return X, y
# evaluate a give model using cross-validation
def evaluate_model(model):
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
return scores
# define dataset
X, y = load_dataset('pima-indians-diabetes.csv')
# define number of features to evaluate
num_features = [i+1 for i in range(X.shape[1])]
# enumerate each number of features
results = list()
for k in num_features:
# create pipeline
model = LogisticRegression(solver='liblinear')
fs = SelectKBest(score_func=f_classif, k=k)
pipeline = Pipeline(steps=[('anova',fs), ('lr', model)])
# evaluate the model
scores = evaluate_model(pipeline)
results.append(scores)
# summarize the results
print('>%d %.3f (%.3f)' % (k, mean(scores), std(scores)))
# plot model performance for comparison
pyplot.boxplot(results, labels=num_features, showmeans=True)
pyplot.show()
Running the example first reports the mean and standard deviation accuracy for each number of selected features.
Your specific results may vary given the stochastic nature of the learning algorithm and evaluation procedure. Try running the example a few times.
In this case, it looks like selecting five and seven features results in roughly the same accuracy.
>1 0.748 (0.048) >2 0.756 (0.042) >3 0.761 (0.044) >4 0.759 (0.042) >5 0.770 (0.041) >6 0.766 (0.042) >7 0.770 (0.042) >8 0.768 (0.040)
Box and whisker plots are created side-by-side showing the trend of increasing mean accuracy with the number of selected features to five features, after which it may become less stable.
Selecting five features might be an appropriate configuration in this case.
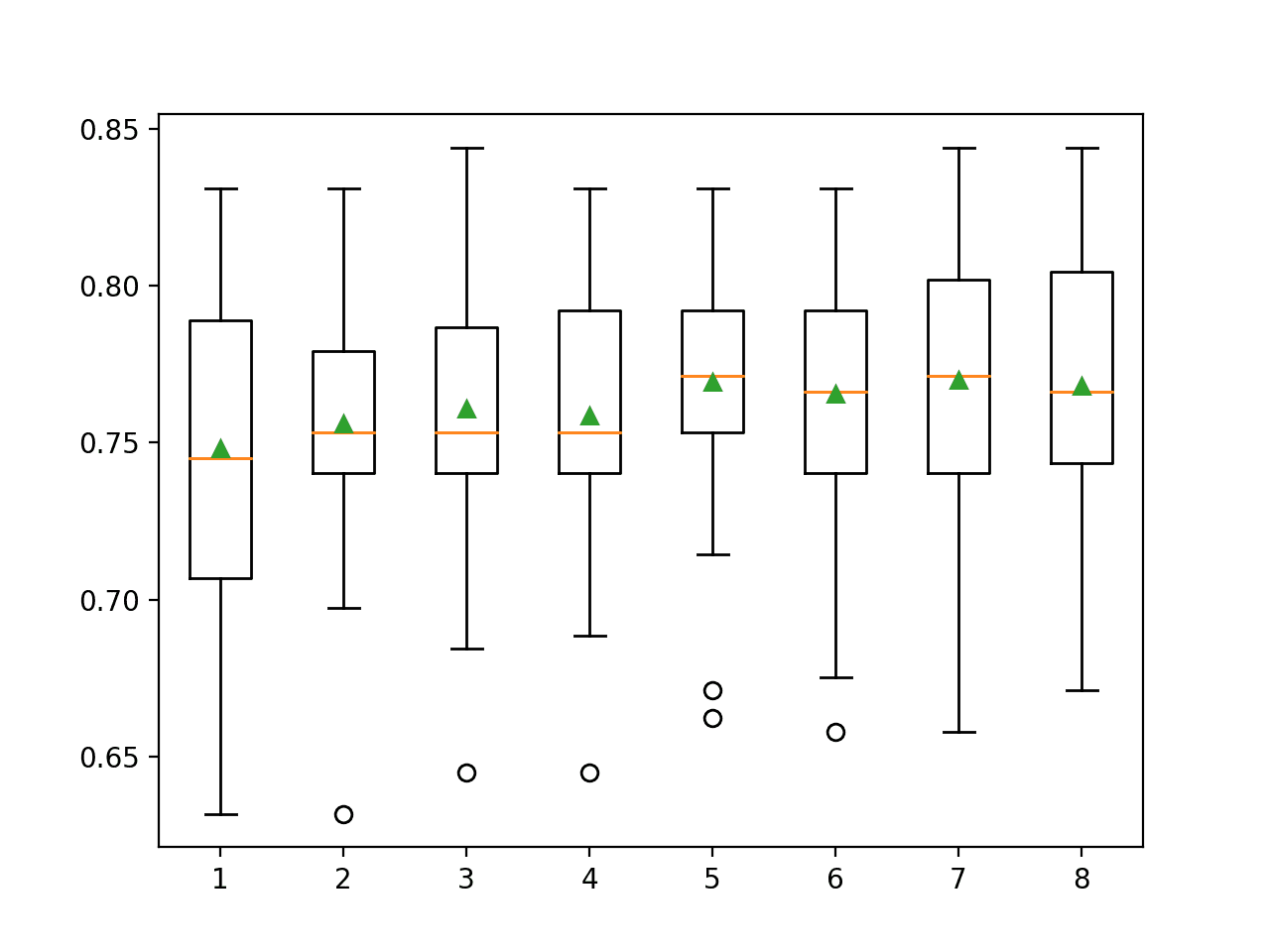
Box and Whisker Plots of Classification Accuracy for Each Number of Selected Features Using ANOVA f-test
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Tutorials
- How to Choose a Feature Selection Method For Machine Learning
- How to Perform Feature Selection with Categorical Data
- What Is Information Gain and Mutual Information for Machine Learning
Books
Papers
APIs
- Feature selection, Scikit-Learn User Guide.
- sklearn.feature_selection.f_classif API.
- sklearn.feature_selection.mutual_info_classif API.
- sklearn.feature_selection.SelectKBest API.
Articles
Datasets
- Diabetes Dataset (pima-indians-diabetes.csv)
- Diabetes Dataset Description (pima-indians-diabetes.names)
Summary
In this tutorial, you discovered how to perform feature selection with numerical input data for classification.
Specifically, you learned:
- The diabetes predictive modeling problem with numerical inputs and binary classification target variables.
- How to evaluate the importance of numerical features using the ANOVA f-test and mutual information statistics.
- How to perform feature selection for numerical data when fitting and evaluating a classification model.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post How to Perform Feature Selection With Numerical Input Data appeared first on Machine Learning Mastery.
