Author: Jason Brownlee
Many machine learning algorithms perform better when numerical input variables are scaled to a standard range.
This includes algorithms that use a weighted sum of the input, like linear regression, and algorithms that use distance measures, like k-nearest neighbors.
The two most popular techniques for scaling numerical data prior to modeling are normalization and standardization. Normalization scales each input variable separately to the range 0-1, which is the range for floating-point values where we have the most precision. Standardization scales each input variable separately by subtracting the mean (called centering) and dividing by the standard deviation to shift the distribution to have a mean of zero and a standard deviation of one.
In this tutorial, you will discover how to use scaler transforms to standardize and normalize numerical input variables for classification and regression.
After completing this tutorial, you will know:
- Data scaling is a recommended pre-processing step when working with many machine learning algorithms.
- Data scaling can be achieved by normalizing or standardizing real-valued input and output variables.
- How to apply standardization and normalization to improve the performance of predictive modeling algorithms.
Let’s get started.

How to Use StandardScaler and MinMaxScaler Transforms
Photo by Marco Verch, some rights reserved.
Tutorial Overview
This tutorial is divided into six parts; they are:
- The Scale of Your Data Matters
- Numerical Data Scaling Methods
- Data Normalization
- Data Standardization
- Sonar Dataset
- MinMaxScaler Transform
- StandardScaler Transform
- Common Questions
The Scale of Your Data Matters
Machine learning models learn a mapping from input variables to an output variable.
As such, the scale and distribution of the data drawn from the domain may be different for each variable.
Input variables may have different units (e.g. feet, kilometers, and hours) that, in turn, may mean the variables have different scales.
Differences in the scales across input variables may increase the difficulty of the problem being modeled. An example of this is that large input values (e.g. a spread of hundreds or thousands of units) can result in a model that learns large weight values. A model with large weight values is often unstable, meaning that it may suffer from poor performance during learning and sensitivity to input values resulting in higher generalization error.
One of the most common forms of pre-processing consists of a simple linear rescaling of the input variables.
— Page 298, Neural Networks for Pattern Recognition, 1995.
This difference in scale for input variables does not affect all machine learning algorithms.
For example, algorithms that fit a model that use a weighted sum of input variables are affected, such as linear regression, logistic regression, and artificial neural networks (deep learning).
For example, when the distance or dot products between predictors are used (such as K-nearest neighbors or support vector machines) or when the variables are required to be a common scale in order to apply a penalty, a standardization procedure is essential.
— Page 124, Feature Engineering and Selection, 2019.
Also, algorithms that use distance measures between examples or exemplars are affected, such as k-nearest neighbors and support vector machines. There are also algorithms that are unaffected by the scale of numerical input variables, most notably decision trees and ensembles of trees, like random forest.
Different attributes are measured on different scales, so if the Euclidean distance formula were used directly, the effect of some attributes might be completely dwarfed by others that had larger scales of measurement. Consequently, it is usual to normalize all attribute values …
— Page 145, Data Mining: Practical Machine Learning Tools and Techniques, 2016.
It can also be a good idea to scale the target variable for regression predictive modeling problems to make the problem easier to learn, most notably in the case of neural network models. A target variable with a large spread of values, in turn, may result in large error gradient values causing weight values to change dramatically, making the learning process unstable.
Scaling input and output variables is a critical step in using neural network models.
In practice, it is nearly always advantageous to apply pre-processing transformations to the input data before it is presented to a network. Similarly, the outputs of the network are often post-processed to give the required output values.
— Page 296, Neural Networks for Pattern Recognition, 1995.
Numerical Data Scaling Methods
Both normalization and standardization can be achieved using the scikit-learn library.
Let’s take a closer look at each in turn.
Data Normalization
Normalization is a rescaling of the data from the original range so that all values are within the new range of 0 and 1.
Normalization requires that you know or are able to accurately estimate the minimum and maximum observable values. You may be able to estimate these values from your available data.
Attributes are often normalized to lie in a fixed range — usually from zero to one—by dividing all values by the maximum value encountered or by subtracting the minimum value and dividing by the range between the maximum and minimum values.
— Page 61, Data Mining: Practical Machine Learning Tools and Techniques, 2016.
A value is normalized as follows:
- y = (x – min) / (max – min)
Where the minimum and maximum values pertain to the value x being normalized.
For example, for a dataset, we could guesstimate the min and max observable values as 30 and -10. We can then normalize any value, like 18.8, as follows:
- y = (x – min) / (max – min)
- y = (18.8 – (-10)) / (30 – (-10))
- y = 28.8 / 40
- y = 0.72
You can see that if an x value is provided that is outside the bounds of the minimum and maximum values, the resulting value will not be in the range of 0 and 1. You could check for these observations prior to making predictions and either remove them from the dataset or limit them to the pre-defined maximum or minimum values.
You can normalize your dataset using the scikit-learn object MinMaxScaler.
Good practice usage with the MinMaxScaler and other scaling techniques is as follows:
- Fit the scaler using available training data. For normalization, this means the training data will be used to estimate the minimum and maximum observable values. This is done by calling the fit() function.
- Apply the scale to training data. This means you can use the normalized data to train your model. This is done by calling the transform() function.
- Apply the scale to data going forward. This means you can prepare new data in the future on which you want to make predictions.
The default scale for the MinMaxScaler is to rescale variables into the range [0,1], although a preferred scale can be specified via the “feature_range” argument and specify a tuple, including the min and the max for all variables.
We can demonstrate the usage of this class by converting two variables to a range 0-to-1, the default range for normalization. The first variable has values between about 4 and 100, the second has values between about 0.1 and 0.001.
The complete example is listed below.
# example of a normalization from numpy import asarray from sklearn.preprocessing import MinMaxScaler # define data data = asarray([[100, 0.001], [8, 0.05], [50, 0.005], [88, 0.07], [4, 0.1]]) print(data) # define min max scaler scaler = MinMaxScaler() # transform data scaled = scaler.fit_transform(data) print(scaled)
Running the example first reports the raw dataset, showing 2 columns with 4 rows. The values are in scientific notation which can be hard to read if you’re not used to it.
Next, the scaler is defined, fit on the whole dataset and then used to create a transformed version of the dataset with each column normalized independently. We can see that the largest raw value for each column now has the value 1.0 and the smallest value for each column now has the value 0.0.
[[1.0e+02 1.0e-03] [8.0e+00 5.0e-02] [5.0e+01 5.0e-03] [8.8e+01 7.0e-02] [4.0e+00 1.0e-01]] [[1. 0. ] [0.04166667 0.49494949] [0.47916667 0.04040404] [0.875 0.6969697 ] [0. 1. ]]
Now that we are familiar with normalization, let’s take a closer look at standardization.
Data Standardization
Standardizing a dataset involves rescaling the distribution of values so that the mean of observed values is 0 and the standard deviation is 1.
This can be thought of as subtracting the mean value or centering the data.
Like normalization, standardization can be useful, and even required in some machine learning algorithms when your data has input values with differing scales.
Standardization assumes that your observations fit a Gaussian distribution (bell curve) with a well-behaved mean and standard deviation. You can still standardize your data if this expectation is not met, but you may not get reliable results.
Another […] technique is to calculate the statistical mean and standard deviation of the attribute values, subtract the mean from each value, and divide the result by the standard deviation. This process is called standardizing a statistical variable and results in a set of values whose mean is zero and standard deviation is one.
— Page 61, Data Mining: Practical Machine Learning Tools and Techniques, 2016.
Standardization requires that you know or are able to accurately estimate the mean and standard deviation of observable values. You may be able to estimate these values from your training data, not the entire dataset.
Again, it is emphasized that the statistics required for the transformation (e.g., the mean) are estimated from the training set and are applied to all data sets (e.g., the test set or new samples).
— Page 124, Feature Engineering and Selection, 2019.
Subtracting the mean from the data is called centering, whereas dividing by the standard deviation is called scaling. As such, the method is sometime called “center scaling“.
The most straightforward and common data transformation is to center scale the predictor variables. To center a predictor variable, the average predictor value is subtracted from all the values. As a result of centering, the predictor has a zero mean. Similarly, to scale the data, each value of the predictor variable is divided by its standard deviation. Scaling the data coerce the values to have a common standard deviation of one.
— Page 30, Applied Predictive Modeling, 2013.
A value is standardized as follows:
- y = (x – mean) / standard_deviation
Where the mean is calculated as:
- mean = sum(x) / count(x)
And the standard_deviation is calculated as:
- standard_deviation = sqrt( sum( (x – mean)^2 ) / count(x))
We can guesstimate a mean of 10.0 and a standard deviation of about 5.0. Using these values, we can standardize the first value of 20.7 as follows:
- y = (x – mean) / standard_deviation
- y = (20.7 – 10) / 5
- y = (10.7) / 5
- y = 2.14
The mean and standard deviation estimates of a dataset can be more robust to new data than the minimum and maximum.
You can standardize your dataset using the scikit-learn object StandardScaler.
We can demonstrate the usage of this class by converting two variables to a range 0-to-1 defined in the previous section. We will use the default configuration that will both center and scale the values in each column, e.g. full standardization.
The complete example is listed below.
# example of a standardization from numpy import asarray from sklearn.preprocessing import StandardScaler # define data data = asarray([[100, 0.001], [8, 0.05], [50, 0.005], [88, 0.07], [4, 0.1]]) print(data) # define standard scaler scaler = StandardScaler() # transform data scaled = scaler.fit_transform(data) print(scaled)
Running the example first reports the raw dataset, showing 2 columns with 4 rows as before.
Next, the scaler is defined, fit on the whole dataset and then used to create a transformed version of the dataset with each column standardized independently. We can see that the mean value in each column is assigned a value of 0.0 if present and the values are centered around 0.0 with values both positive and negative.
[[1.0e+02 1.0e-03] [8.0e+00 5.0e-02] [5.0e+01 5.0e-03] [8.8e+01 7.0e-02] [4.0e+00 1.0e-01]] [[ 1.26398112 -1.16389967] [-1.06174414 0.12639634] [ 0. -1.05856939] [ 0.96062565 0.65304778] [-1.16286263 1.44302493]]
Next, we can introduce a real dataset that provides the basis for applying normalization and standardization transforms as a part of modeling.
Sonar Dataset
The sonar dataset is a standard machine learning dataset for binary classification.
It involves 60 real-valued inputs and a two-class target variable. There are 208 examples in the dataset and the classes are reasonably balanced.
A baseline classification algorithm can achieve a classification accuracy of about 53.4 percent using repeated stratified 10-fold cross-validation. Top performance on this dataset is about 88 percent using repeated stratified 10-fold cross-validation.
The dataset describes radar returns of rocks or simulated mines.
You can learn more about the dataset from here:
No need to download the dataset; we will download it automatically from our worked examples.
First, let’s load and summarize the dataset. The complete example is listed below.
# load and summarize the sonar dataset from pandas import read_csv from pandas.plotting import scatter_matrix from matplotlib import pyplot # Load dataset url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv" dataset = read_csv(url, header=None) # summarize the shape of the dataset print(dataset.shape) # summarize each variable print(dataset.describe()) # histograms of the variables dataset.hist() pyplot.show()
Running the example first summarizes the shape of the loaded dataset.
This confirms the 60 input variables, one output variable, and 208 rows of data.
A statistical summary of the input variables is provided showing that values are numeric and range approximately from 0 to 1.
(208, 61)
0 1 2 ... 57 58 59
count 208.000000 208.000000 208.000000 ... 208.000000 208.000000 208.000000
mean 0.029164 0.038437 0.043832 ... 0.007949 0.007941 0.006507
std 0.022991 0.032960 0.038428 ... 0.006470 0.006181 0.005031
min 0.001500 0.000600 0.001500 ... 0.000300 0.000100 0.000600
25% 0.013350 0.016450 0.018950 ... 0.003600 0.003675 0.003100
50% 0.022800 0.030800 0.034300 ... 0.005800 0.006400 0.005300
75% 0.035550 0.047950 0.057950 ... 0.010350 0.010325 0.008525
max 0.137100 0.233900 0.305900 ... 0.044000 0.036400 0.043900
[8 rows x 60 columns]
Finally, a histogram is created for each input variable.
If we ignore the clutter of the plots and focus on the histograms themselves, we can see that many variables have a skewed distribution.
The dataset provides a good candidate for using scaler transforms as the variables have differing minimum and maximum values, as well as different data distributions.
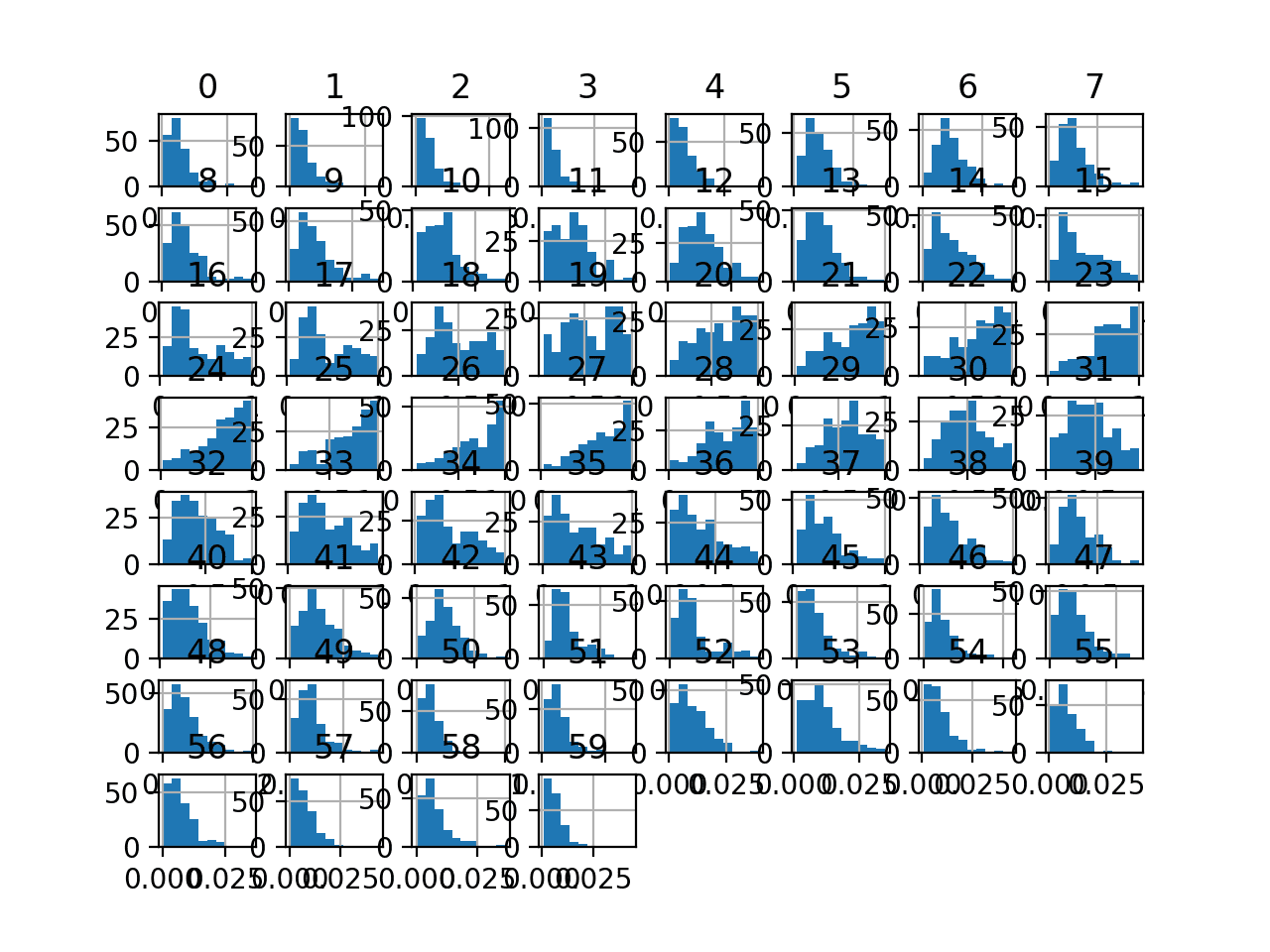
Histogram Plots of Input Variables for the Sonar Binary Classification Dataset
Next, let’s fit and evaluate a machine learning model on the raw dataset.
We will use a k-nearest neighbor algorithm with default hyperparameters and evaluate it using repeated stratified k-fold cross-validation. The complete example is listed below.
# evaluate knn on the raw sonar dataset
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import LabelEncoder
from matplotlib import pyplot
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
# define and configure the model
model = KNeighborsClassifier()
# evaluate the model
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(model, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report model performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example evaluates a KNN model on the raw sonar dataset.
We can see that the model achieved a mean classification accuracy of about 79.7 percent, showing that it has skill (better than 53.4 percent) and is in the ball-park of good performance (88 percent).
Accuracy: 0.797 (0.073)
Next, let’s explore a scaling transform of the dataset.
MinMaxScaler Transform
We can apply the MinMaxScaler to the Sonar dataset directly to normalize the input variables.
We will use the default configuration and scale values to the range 0 and 1. First, a MinMaxScaler instance is defined with default hyperparameters. Once defined, we can call the fit_transform() function and pass it to our dataset to create a transformed version of our dataset.
... # perform a robust scaler transform of the dataset trans = MinMaxScaler() data = trans.fit_transform(data)
Let’s try it on our sonar dataset.
The complete example of creating a MinMaxScaler transform of the sonar dataset and plotting histograms of the result is listed below.
# visualize a minmax scaler transform of the sonar dataset from pandas import read_csv from pandas import DataFrame from pandas.plotting import scatter_matrix from sklearn.preprocessing import MinMaxScaler from matplotlib import pyplot # load dataset url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv" dataset = read_csv(url, header=None) # retrieve just the numeric input values data = dataset.values[:, :-1] # perform a robust scaler transform of the dataset trans = MinMaxScaler() data = trans.fit_transform(data) # convert the array back to a dataframe dataset = DataFrame(data) # summarize print(dataset.describe()) # histograms of the variables dataset.hist() pyplot.show()
Running the example first reports a summary of each input variable.
We can see that the distributions have been adjusted and that the minimum and maximum values for each variable are now a crisp 0.0 and 1.0 respectively.
0 1 2 ... 57 58 59 count 208.000000 208.000000 208.000000 ... 208.000000 208.000000 208.000000 mean 0.204011 0.162180 0.139068 ... 0.175035 0.216015 0.136425 std 0.169550 0.141277 0.126242 ... 0.148051 0.170286 0.116190 min 0.000000 0.000000 0.000000 ... 0.000000 0.000000 0.000000 25% 0.087389 0.067938 0.057326 ... 0.075515 0.098485 0.057737 50% 0.157080 0.129447 0.107753 ... 0.125858 0.173554 0.108545 75% 0.251106 0.202958 0.185447 ... 0.229977 0.281680 0.183025 max 1.000000 1.000000 1.000000 ... 1.000000 1.000000 1.000000 [8 rows x 60 columns]
Histogram plots of the variables are created, although the distributions don’t look much different from their original distributions seen in the previous section.
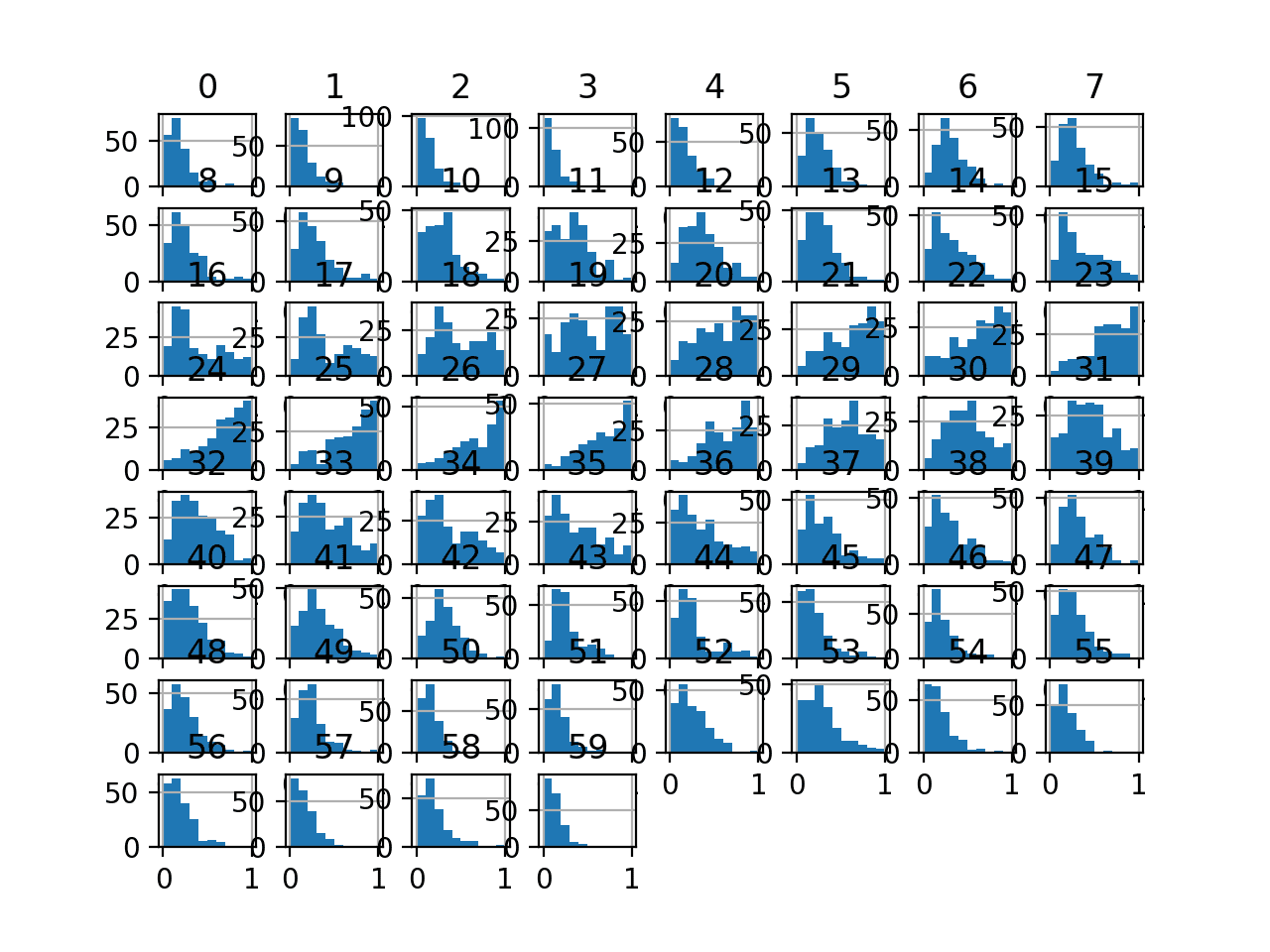
Histogram Plots of MinMaxScaler Transformed Input Variables for the Sonar Dataset
Next, let’s evaluate the same KNN model as the previous section, but in this case, on a MinMaxScaler transform of the dataset.
The complete example is listed below.
# evaluate knn on the sonar dataset with minmax scaler transform
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import MinMaxScaler
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
# define the pipeline
trans = MinMaxScaler()
model = KNeighborsClassifier()
pipeline = Pipeline(steps=[('t', trans), ('m', model)])
# evaluate the pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(pipeline, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report pipeline performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example, we can see that the MinMaxScaler transform results in a lift in performance from 79.7 percent accuracy without the transform to about 81.3 percent with the transform.
Accuracy: 0.813 (0.085)
Next, let’s explore the effect of standardizing the input variables.
StandardScaler Transform
We can apply the StandardScaler to the Sonar dataset directly to standardize the input variables.
We will use the default configuration and scale values to subtract the mean to center them on 0.0 and divide by the standard deviation to give the standard deviation of 1.0. First, a StandardScaler instance is defined with default hyperparameters.
Once defined, we can call the fit_transform() function and pass it to our dataset to create a transformed version of our dataset.
... # perform a robust scaler transform of the dataset trans = StandardScaler() data = trans.fit_transform(data)
Let’s try it on our sonar dataset.
The complete example of creating a StandardScaler transform of the sonar dataset and plotting histograms of the results is listed below.
# visualize a standard scaler transform of the sonar dataset from pandas import read_csv from pandas import DataFrame from pandas.plotting import scatter_matrix from sklearn.preprocessing import StandardScaler from matplotlib import pyplot # load dataset url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv" dataset = read_csv(url, header=None) # retrieve just the numeric input values data = dataset.values[:, :-1] # perform a robust scaler transform of the dataset trans = StandardScaler() data = trans.fit_transform(data) # convert the array back to a dataframe dataset = DataFrame(data) # summarize print(dataset.describe()) # histograms of the variables dataset.hist() pyplot.show()
Running the example first reports a summary of each input variable.
We can see that the distributions have been adjusted and that the mean is a very small number close to zero and the standard deviation is very close to 1.0 for each variable.
0 1 ... 58 59 count 2.080000e+02 2.080000e+02 ... 2.080000e+02 2.080000e+02 mean -4.190024e-17 1.663333e-16 ... 1.283695e-16 3.149190e-17 std 1.002413e+00 1.002413e+00 ... 1.002413e+00 1.002413e+00 min -1.206158e+00 -1.150725e+00 ... -1.271603e+00 -1.176985e+00 25% -6.894939e-01 -6.686781e-01 ... -6.918580e-01 -6.788714e-01 50% -2.774703e-01 -2.322506e-01 ... -2.499546e-01 -2.405314e-01 75% 2.784345e-01 2.893335e-01 ... 3.865486e-01 4.020352e-01 max 4.706053e+00 5.944643e+00 ... 4.615037e+00 7.450343e+00 [8 rows x 60 columns]
Histogram plots of the variables are created, although the distributions don’t look much different from their original distributions seen in the previous section other than their scale on the x-axis.
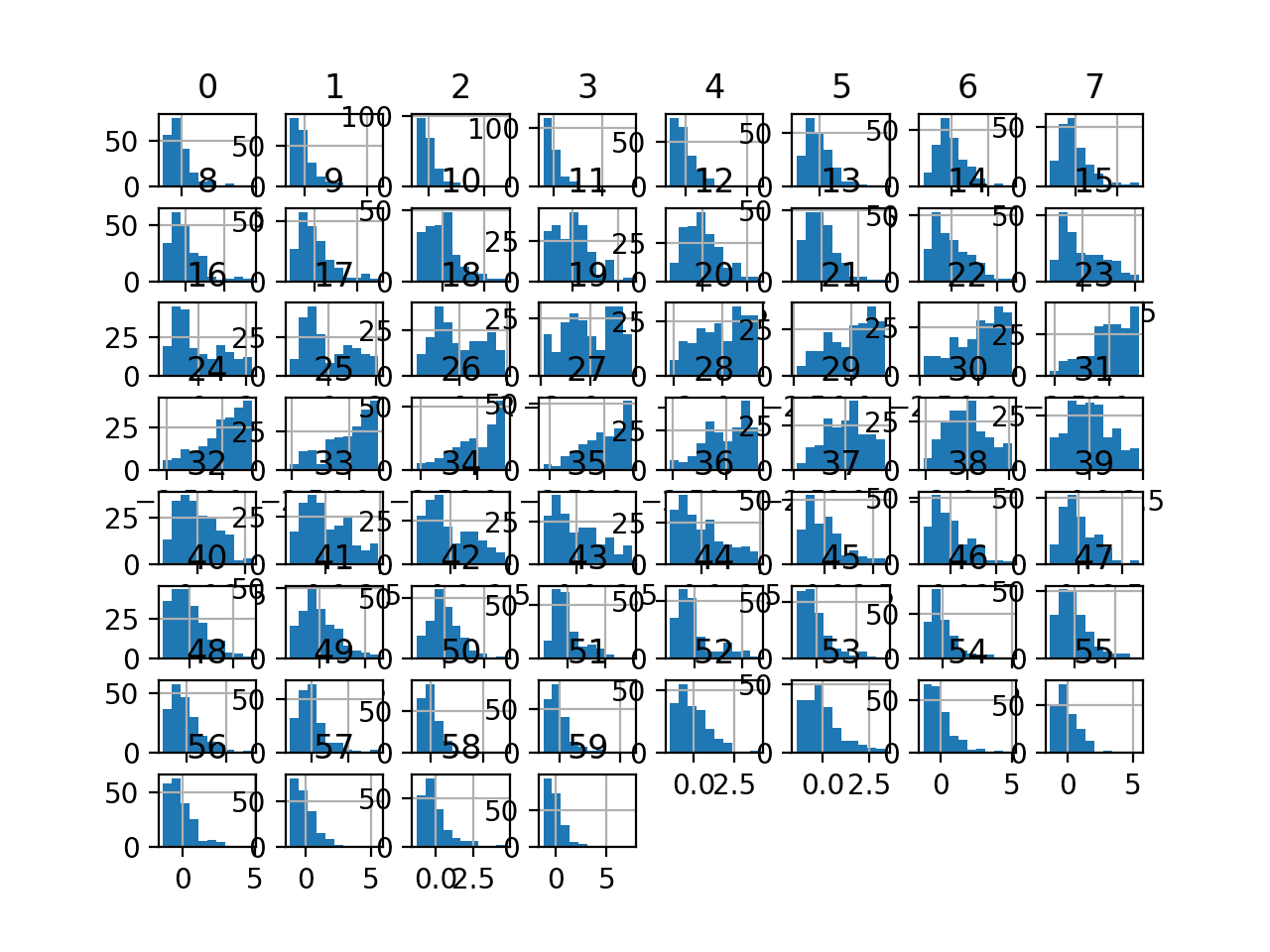
Histogram Plots of StandardScaler Transformed Input Variables for the Sonar Dataset
Next, let’s evaluate the same KNN model as the previous section, but in this case, on a StandardScaler transform of the dataset.
The complete example is listed below.
# evaluate knn on the sonar dataset with standard scaler transform
from numpy import mean
from numpy import std
from pandas import read_csv
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
from matplotlib import pyplot
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/sonar.csv"
dataset = read_csv(url, header=None)
data = dataset.values
# separate into input and output columns
X, y = data[:, :-1], data[:, -1]
# ensure inputs are floats and output is an integer label
X = X.astype('float32')
y = LabelEncoder().fit_transform(y.astype('str'))
# define the pipeline
trans = StandardScaler()
model = KNeighborsClassifier()
pipeline = Pipeline(steps=[('t', trans), ('m', model)])
# evaluate the pipeline
cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
n_scores = cross_val_score(pipeline, X, y, scoring='accuracy', cv=cv, n_jobs=-1, error_score='raise')
# report pipeline performance
print('Accuracy: %.3f (%.3f)' % (mean(n_scores), std(n_scores)))
Running the example, we can see that the StandardScaler transform results in a lift in performance from 79.7 percent accuracy without the transform to about 81.0 percent with the transform, although slightly lower than the result using the MinMaxScaler.
Accuracy: 0.810 (0.080)
Common Questions
This section lists some common questions and answers when scaling numerical data.
Q. Should I Normalize or Standardize?
Whether input variables require scaling depends on the specifics of your problem and of each variable.
You may have a sequence of quantities as inputs, such as prices or temperatures.
If the distribution of the quantity is normal, then it should be standardized, otherwise, the data should be normalized. This applies if the range of quantity values is large (10s, 100s, etc.) or small (0.01, 0.0001).
If the quantity values are small (near 0-1) and the distribution is limited (e.g. standard deviation near 1), then perhaps you can get away with no scaling of the data.
These manipulations are generally used to improve the numerical stability of some calculations. Some models […] benefit from the predictors being on a common scale.
— Pages 30-31, Applied Predictive Modeling, 2013.
Predictive modeling problems can be complex, and it may not be clear how to best scale input data.
If in doubt, normalize the input sequence. If you have the resources, explore modeling with the raw data, standardized data, and normalized data and see if there is a beneficial difference in the performance of the resulting model.
If the input variables are combined linearly, as in an MLP [Multilayer Perceptron], then it is rarely strictly necessary to standardize the inputs, at least in theory. […] However, there are a variety of practical reasons why standardizing the inputs can make training faster and reduce the chances of getting stuck in local optima.
— Should I normalize/standardize/rescale the data? Neural Nets FAQ
Q. Should I Standardize then Normalize?
Standardization can give values that are both positive and negative centered around zero.
It may be desirable to normalize data after it has been standardized.
This might be a good idea of you have a mixture of standardized and normalized variables and wish all input variables to have the same minimum and maximum values as input for a given algorithm, such as an algorithm that calculates distance measures.
Q. But Which is Best?
This is unknowable.
Evaluate models on data prepared with each transform and use the transform or combination of transforms that result in the best performance for your data set on your model.
Q. How Do I Handle Out-of-Bounds Values?
You may normalize your data by calculating the minimum and maximum on the training data.
Later, you may have new data with values smaller or larger than the minimum or maximum respectively.
One simple approach to handling this may be to check for such out-of-bound values and change their values to the known minimum or maximum prior to scaling. Alternately, you may want to estimate the minimum and maximum values used in the normalization manually based on domain knowledge.
Further Reading
This section provides more resources on the topic if you are looking to go deeper.
Tutorials
- How to use Data Scaling Improve Deep Learning Model Stability and Performance
- Rescaling Data for Machine Learning in Python with Scikit-Learn
- 4 Common Machine Learning Data Transforms for Time Series Forecasting
- How to Scale Data for Long Short-Term Memory Networks in Python
- How to Normalize and Standardize Time Series Data in Python
Books
- Neural Networks for Pattern Recognition, 1995.
- Feature Engineering and Selection, 2019.
- Data Mining: Practical Machine Learning Tools and Techniques, 2016.
- Applied Predictive Modeling, 2013.
APIs
Articles
Summary
In this tutorial, you discovered how to use scaler transforms to standardize and normalize numerical input variables for classification and regression.
Specifically, you learned:
- Data scaling is a recommended pre-processing step when working with many machine learning algorithms.
- Data scaling can be achieved by normalizing or standardizing real-valued input and output variables.
- How to apply standardization and normalization to improve the performance of predictive modeling algorithms.
Do you have any questions?
Ask your questions in the comments below and I will do my best to answer.
The post How to Use StandardScaler and MinMaxScaler Transforms in Python appeared first on Machine Learning Mastery.
